Myeloid carcinoma of the colorectum (MeC) is a rare pathological subtype of colorectal cancer, characterized by unique histological and clinical features. It has a rich infiltration of lymphocytes, suggesting the presence of an active immune response. However, previously, comprehensive studies on its immune landscape and the therapeutic effects of immune checkpoint blockade (ICB) have been limited.
iGeneTech Immune Repertoire Assists in Research Sequencing
On December 9, 2024, the team led by Professor Zhang Yanqiao from the Cancer Hospital Affiliated to Harbin Medical University published a research article titled "Multiomics reveals the immunological features and the immune checkpoint blockade potential of colorectal medullary carcinoma" in Clinical Cancer Research. The research team conducted genomic and transcriptomic analyses on 71 patient samples of MeC from the Cancer Hospital Affiliated to Harbin Medical University (HMUCH) and the The Cancer Genome Atlas (TCGA-COAD) database. Combining technologies such as immune cell staining and T cell receptor sequencing, they systematically elaborated on the immune characteristics of MeC, revealed the characteristics of the immune microenvironment of MeC, and explored its potential benefits in immune checkpoint blockade (ICB) therapy. iGeneTech TCR immune repertoire sequencing technology assisted in the part of T cell antigen receptor sequencing for this research.

Sample Source
The research team collected tumor tissue samples of colorectal medullary carcinoma (MeC) and non-medullary carcinoma (nMeC) from the Affiliated Tumor Hospital of Harbin Medical University (HMUCH). After extracting DNA from these samples, appropriate samples were screened out for subsequent T cell receptor sequencing (TCR-seq).
Technical Scheme of the Immunome Repertoire
For the part of T cell receptor sequencing, the TCR detection product of iGeneTech MultipSeq® Human TCR Research Assay, was used for the experiment and analysis.
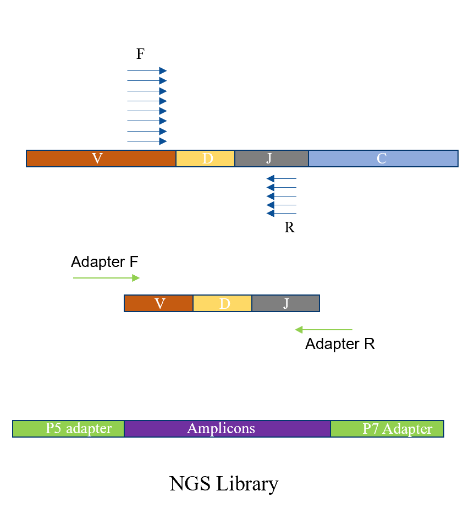
Technical Process
Primers are designed for the V and J regions of the Complementary Determining Region 3 (CDR3) of the TCR. The CDR3 region of the TCR is amplified using multiplex PCR technology, and an adapter sequence is added to construct a library containing the target sequence of the CDR3 region. The constructed library is then subjected to high-depth sequencing on the Illumina NovaSeq 6000 high-throughput sequencing platform.
After the sequencing is completed, the VDJ information of the raw data is analyzed based on the MiXCR software to obtain the sequence information of the TCR chains, providing a data basis for the subsequent analysis of T cell clonality. Computational and visual analysis of the T cell repertoire is carried out.
Key parameters such as the length distribution of CDR3, the number of clones, the number of clonotypes, and the proportion of dominant clones are given special attention. Through these parameters, the clonal ability of immune infiltration is comprehensively evaluated, so as to deeply explore the differences and characteristics of T cell clonality between MeC and nMeC.
Research Results
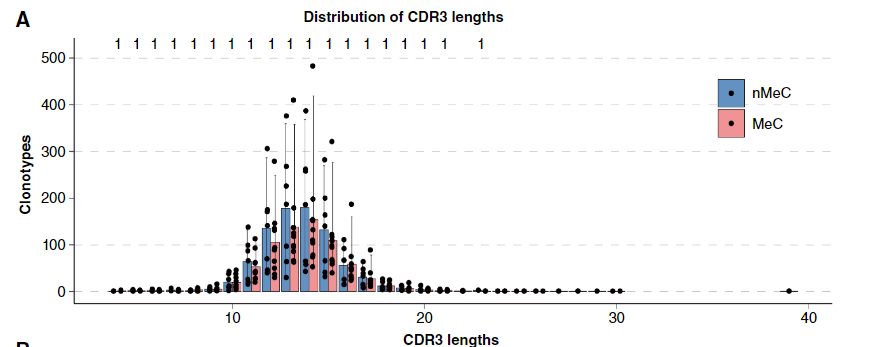
1. Characteristics of CDR3 length: In both MeC and nMeC, the median length of the CDR3 region is 14 amino acids, but the CDR3 region of MeC is slightly longer than that of nMeC. This subtle difference may be of great significance in the process of antigen recognition, suggesting that the antigen recognition characteristics of T cells in MeC may be different from those in nMeC, which provides a clue for further research on the immune response mechanism of MeC.
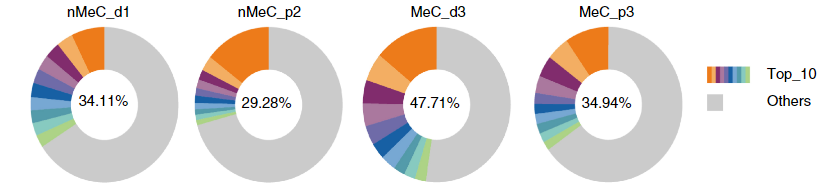
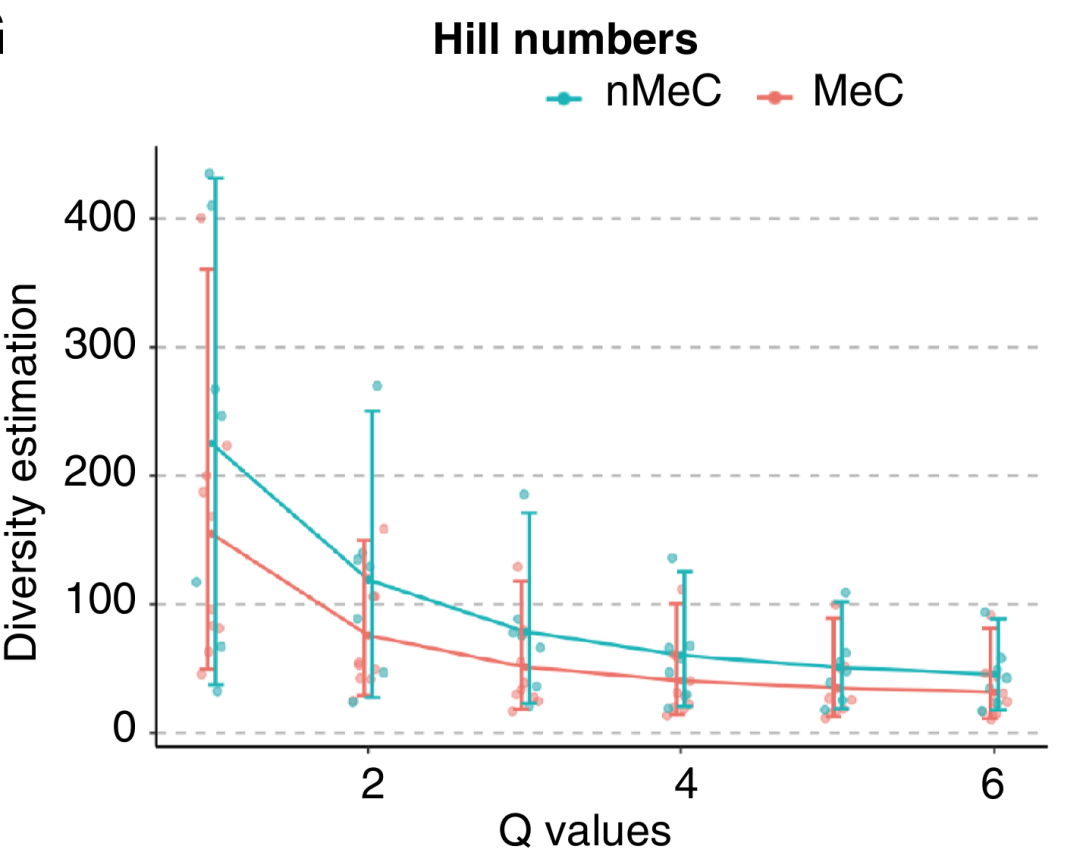
2. Differences in clonotype composition: Through the analysis of the top 10 V-J combinations in the samples, it was found that the top 10 clonotypes accounted for approximately 30% of all sequencing read lengths. Among them, the MeC samples tended to exhibit a higher proportion of the top 10 clonotypes, indicating that the distribution of T cell clonotypes in MeC was relatively concentrated. There may be certain dominant clonotypes playing a key role in the immune response, which forms a clear contrast with nMeC.
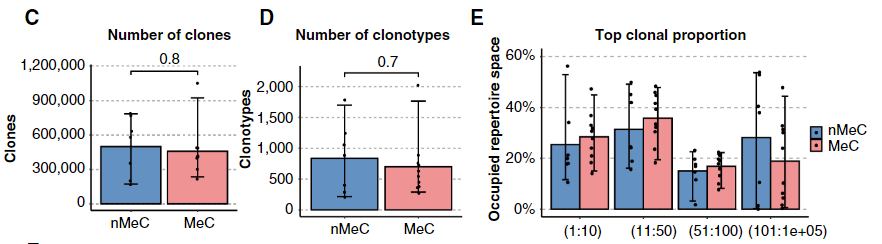
3. Changes in clonality-related indicators: MeC shows a unique trend in clonality-related indicators. The number of clones and the number of clonotypes in MeC are relatively low, while the proportion of dominant clones is relatively high. This indicates that the T cell population in MeC has a stronger clonal expansion ability, that is, a small number of clonotypes have been greatly expanded in the tumor immune microenvironment and dominate the immune response, further reflecting the uniqueness of T cell clonality in MeC.
4. Results of diversity assessment: The diversity of the samples was evaluated using Hill numbers. The results showed that the diversity of MeC was lower than that of nMeC, which once again confirmed the conclusion that the T cell clonality in MeC is higher. The characteristics of T cells with such low diversity and high clonality may be closely related to the mechanisms of tumorigenesis, development, and immune escape of MeC, providing an important basis for a deeper understanding of the immunobiological characteristics of MeC.
Summary
Through a systematic experimental protocol and in-depth data analysis, this study comprehensively revealed the characteristics of T cell clonality in MeC. These results have laid a solid foundation for further exploration of the immune mechanism and immunotherapy response of MeC, and provided a new group of potentially beneficial patients for the immunotherapy of colorectal cancer. In the future, it is possible to further explore its immune mechanism and tumor antigens, and optimize the immunotherapy strategy.
*The TCR product contains α, β, γ and δ chains.
** The BCR product contains IgH, Igκ and Igλ chains.
Immunome repertoire products also for non-human species, such as mice, rats, pigs, etc., all support customization.