Product Introduction
Designed based on MultipSeq® multiplex amplicon sequencing technology, this product targets 226 STR loci in forensic science, including 113 Y-chromosome, 72 autosomal, 41 X-chromosome loci, and sex identification sites. It fully covers all loci specified in GB/T 41009-2021 Forensic Science - Selected Loci and Their Data Structures for DNA Databases and GB/T 44393-2024 Nomenclature Rules for Sequence Polymorphic STR Alleles. This product is applicable for Chinese DNA database information storage, individual identification, scientific research, and more.
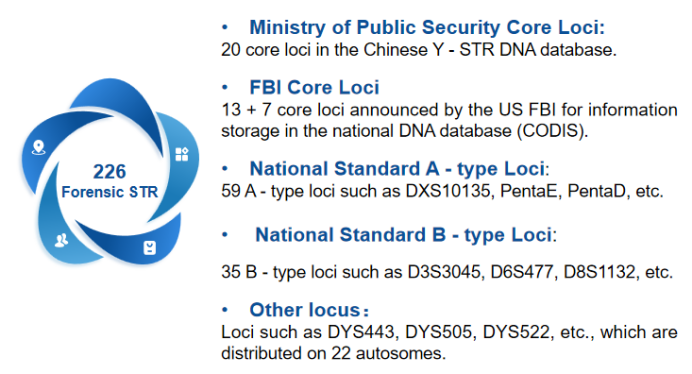
Note: The above is the classification of STR loci, and there will be intersections in some classifications.
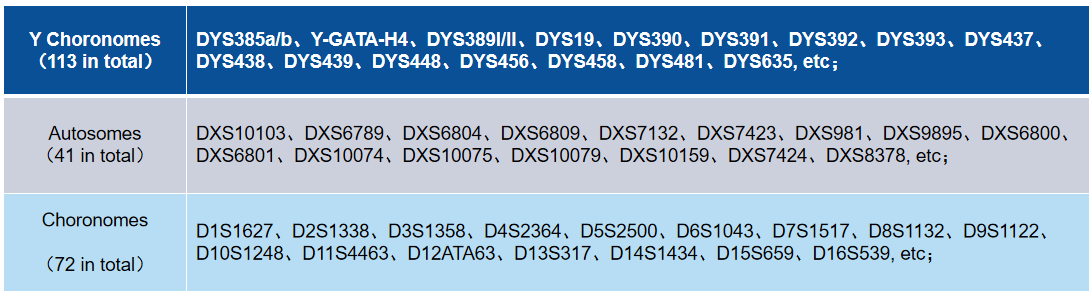
Table 1. Selected STR Loci in 226 STR Research Panel.
STR Analysis Workflow
Utilizing the STR analysis workflow independently developed by iGeneTech, this process can accurately genotype STR sequencing data. The analysis results include information such as loci, allele length, read count and proportion, sequence-based genotyping results, GB-standard length-based genotyping results, and core sequence, enabling comprehensive locus genotyping information.
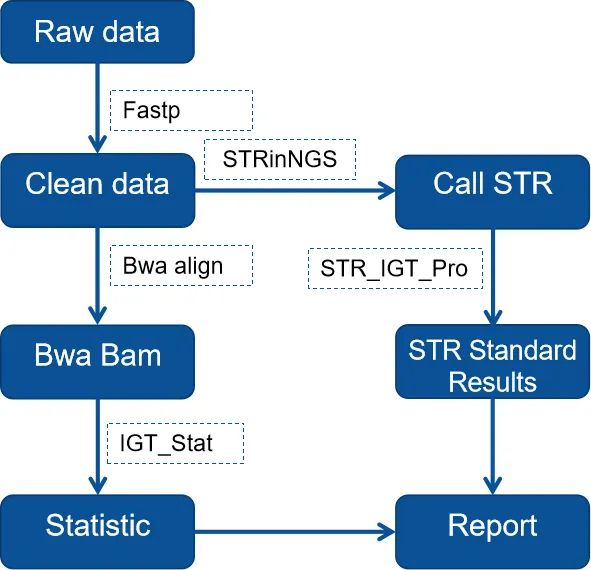
Figure 1. Schematic diagram of iGeneTech STR analysis process
Product Advantages
High Efficiency & Precision: Single-tube amplification enables simultaneous detection of 220+ STR loci in one run.
Low Starting Amount: Requires as little as 125 pg DNA input, supporting direct amplification from blood cards and saliva samples.
Strong Inhibitor Tolerance: Resistant to common inhibitors including 0.05 μg/mL heparin sodium, 100 μM indigo, 100 μM heme, and 100 ng/μL humic acid.
Broad Compatibility: Works with Illumina, MGI, GeneMind, Salus, and other sequencing platforms, supporting read lengths such as PE250, PE300, and SE400.
Flexible Customization: Offers predefined products and supports semi-custom/full-custom solutions for detecting additional genetic markers in a single assay.
End-to-End Solution: Independently developed workflow from sample extraction to data genotyping, delivering CE-consistent length-based genotyping results.
Performance
Data Performance at Different Input Amounts
The 226 STR Research Panel can start with as low as 125 pg of DNA input, and exhibits good performance across all library preparation data metrics starting from 125 pg.
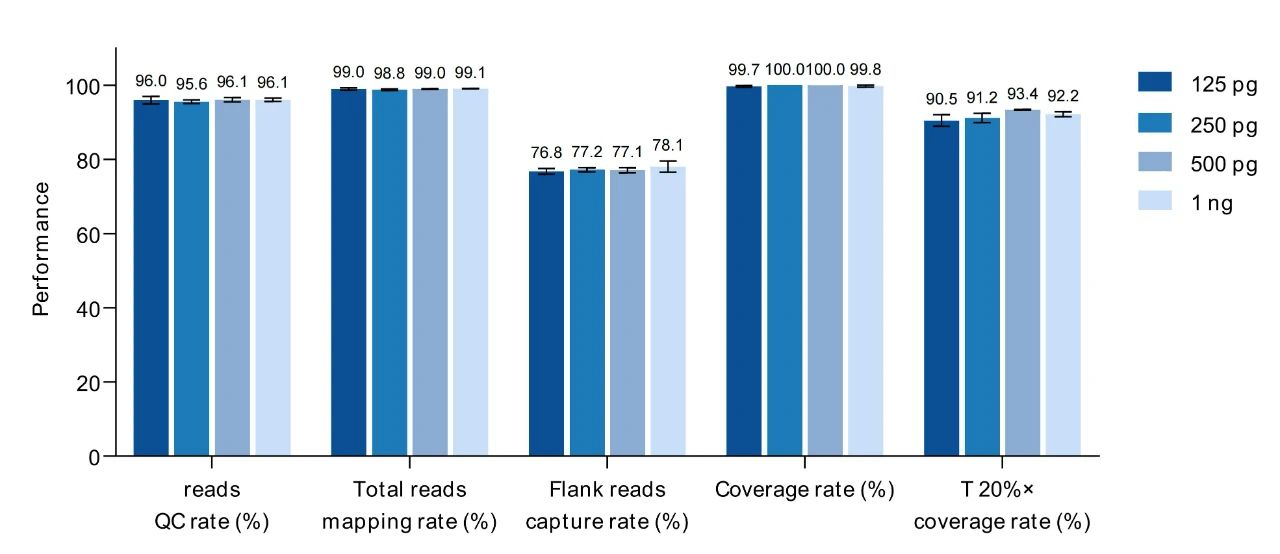
Figure 2. Data Performance of MultipSeq® 226 STR Research Assay (for Illumina TS) at Different Input Amounts. Samples used were 2800M standard reference material with input amounts of 125 pg, 250 pg, 500 pg, and 1 ng. Sequencing was performed on an Illumina NovaSeq 6000 using PE250 chemistry, generating ~0.8M reads with an average sequencing depth of ~2500×.
Detection results of the standard reference material
Based on the test results with input amounts ranging from 125 pg to 1 ng, relatively stable and consistent detection can be achieved, and the locus detection rate can reach over 99%.
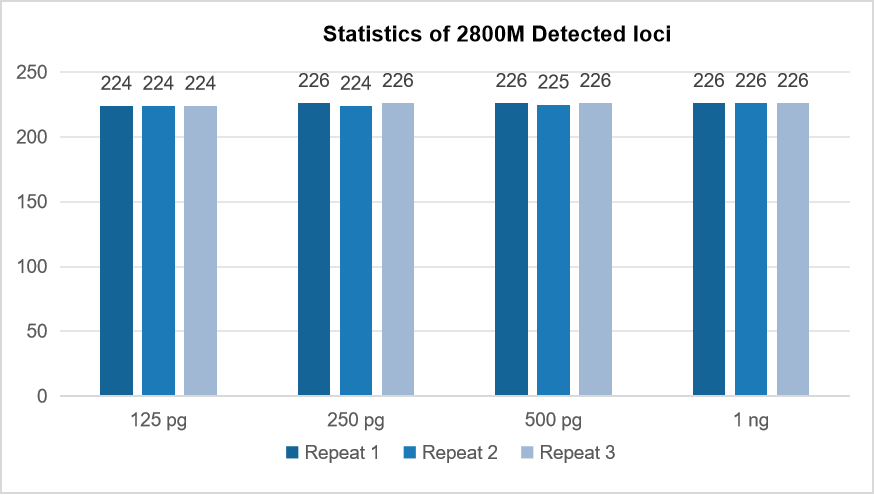
Figure 3. Statistics of the number of STR loci detected in MultipSeq ® 226 STR Research Assay (for Illumina TS) for 2800M standards
Genotyping Results Demonstration
Testing was performed using 2800M standard reference materials, yielding consistent results with known genotypes.
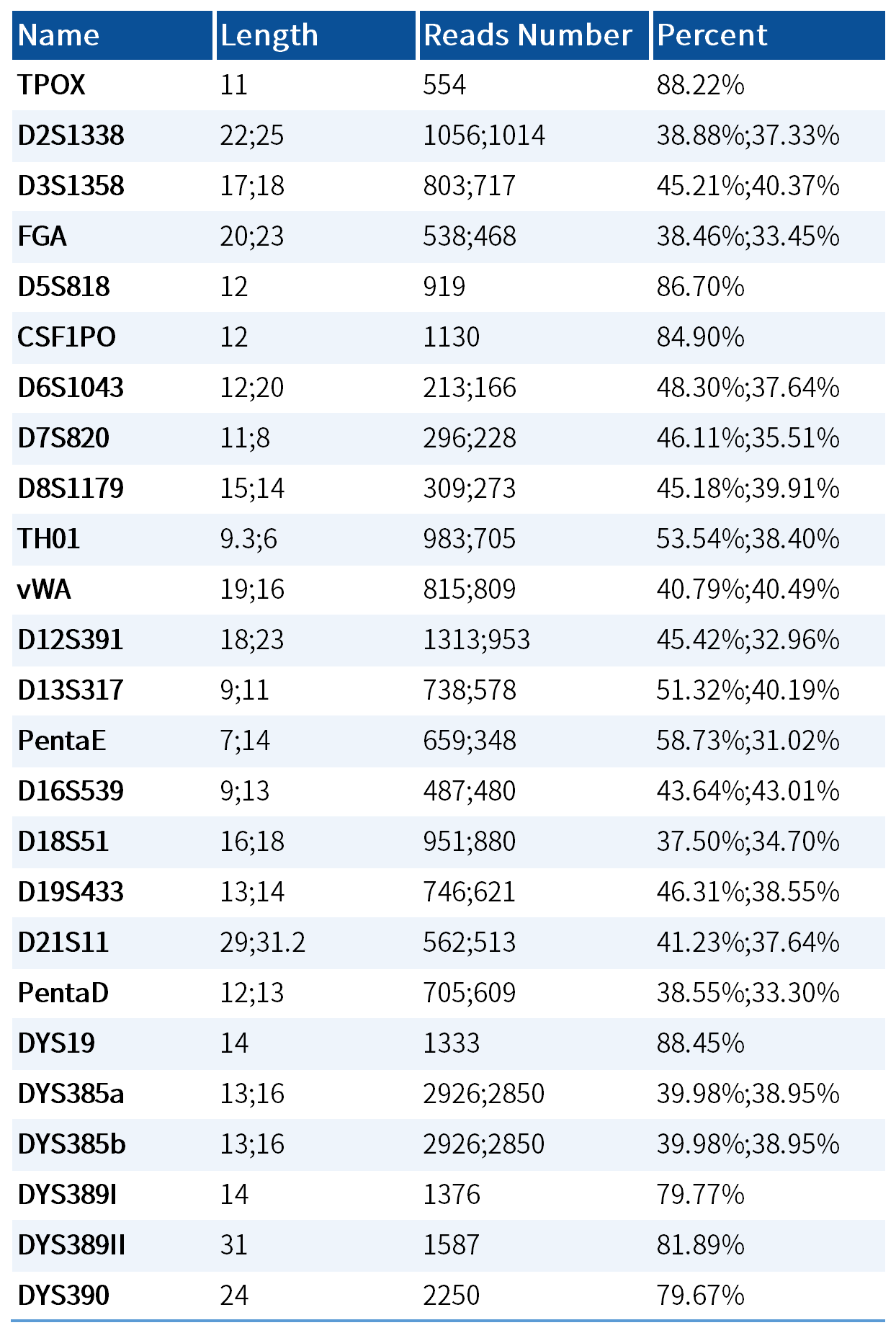
Table 2. 2800M standard partial locus typing results display
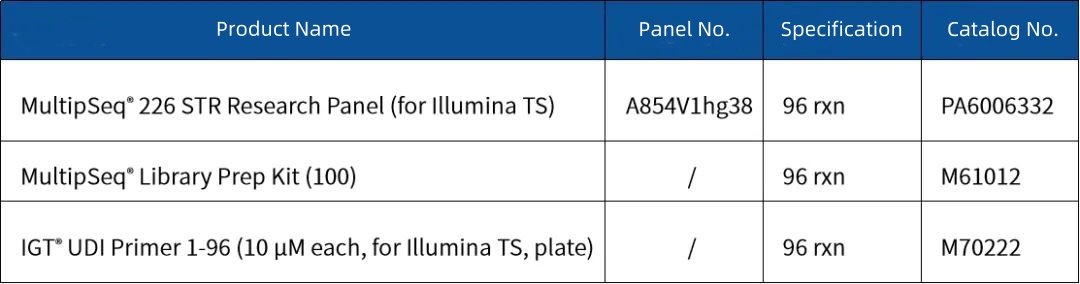
Order Information