Repetitive DNA sequences such as Alu and Kpn I are widely distributed in the human genome. During liquid-phase hybridization capture workflows, these repetitive DNA sequences cause non-specific probe binding and inter-library interactions, thereby reducing panel capture efficiency. Human Block can bind to repetitive sequences in the human genome, blocking these non-specific binding sites and improving panel on-target rates.
The commonly used Human Block in the market is Cot-1 DNA. The preparation steps of Cot-1 DNA involve genomic DNA fragmentation, denaturation, and annealing to enrich repetitive DNA¹. Literature reports that adding Cot-1 DNA to hybridization systems significantly enhances target region capture efficiency².
In liquid-phase hybridization capture workflows, Human Block is typically used in conjunction with Universal Blocking Oligo. Human Block blocks repetitive sequences in the human genome, while Universal blocking oligo blocks adapter sequences, improving targeted capture specificity and data utilization rates.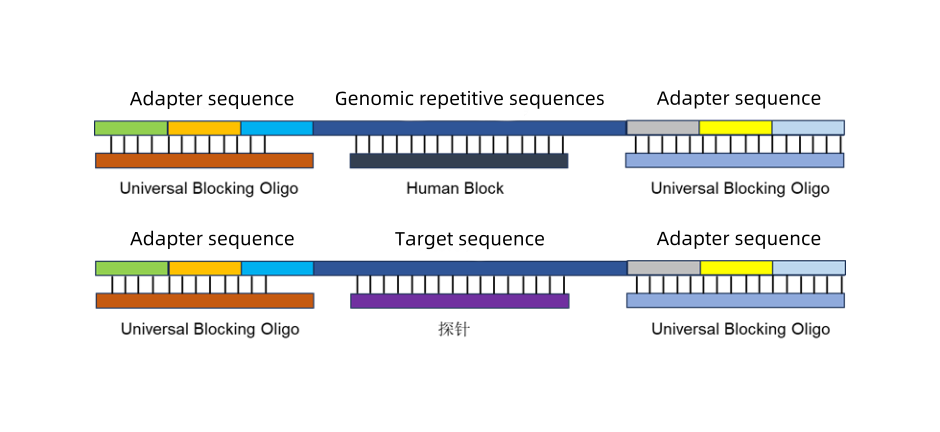
iGeneTech Hyb Human Block
As a reagent supplier specialized in targeted capture, iGeneTech has launched Hyb Human Block, a self-developed and self-produced blocking reagent. Using iGeneTech's proprietary production process, this product can efficiently block repetitive sequences in the human genome during capture workflows. Compared with reagents from other manufacturers in the market, iGeneTech's Hyb Human Block demonstrates higher capture efficiency and better blocking performance:
1. In 2-hour rapid hybridization tests, iGeneTech Hyb Human Block achieved a capture efficiency of 71%, compared to 65% for competitor Human Block. No significant differences were observed in mapping rates or uniformity (Figure 1).
2. In overnight hybridization tests, iGeneTech Hyb Human Block achieved a capture efficiency of 80%, compared to 78% for competitor Human Block. No significant differences were observed in mapping rates or uniformity (Figure 2).
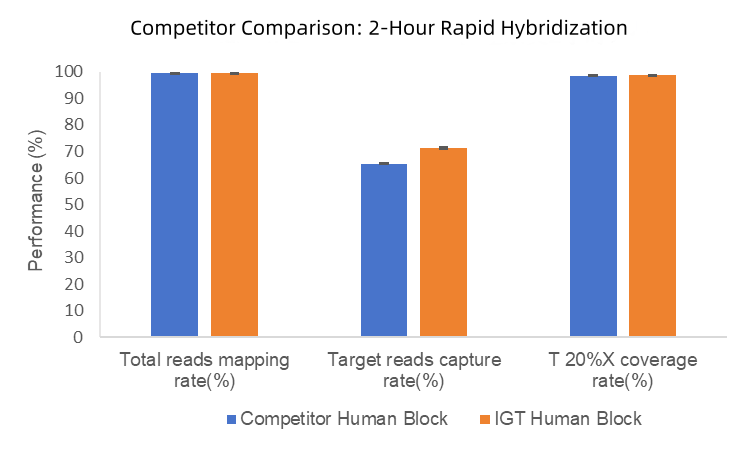
Figure 1. Blocking Performance Comparison Between IGT Human Block and Competitor.
Testing was performed using a 170Kb panel and TargetSeq One v2.0 Capture Kit, with 2-hour rapid hybridization and NovaSeq 6000 platform PE150 sequencing.
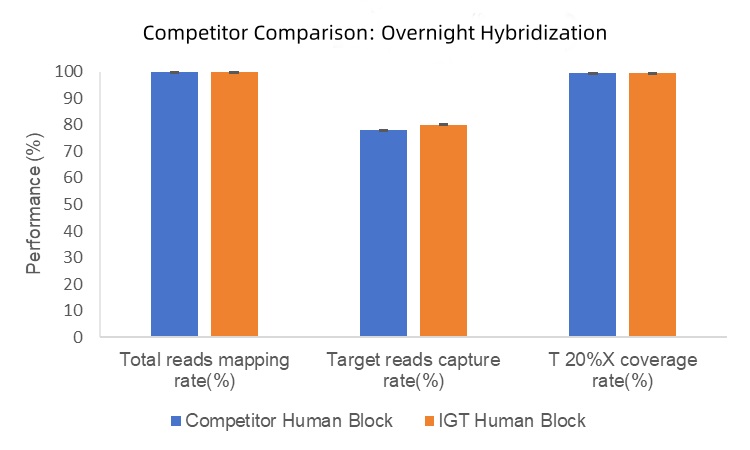
Figure 2. Blocking Performance Comparison Between IGT Human Block and Competitor.
Testing was performed using a 170Kb panel and TargetSeq One v2.0 Capture Kit, with overnight hybridization and NovaSeq 6000 platform PE150 sequencing.
Stability Testing of Hyb Human Block
Three production batches of Hyb Human Block were tested for stability using 2-hour rapid hybridization. Experimental results demonstrate stable performance across batches, with mapping rates >99%, 98% of regions achieving depths exceeding 20% of the average depth, and a capture efficiency coefficient of variation (CV) = 1.2% (Figure 3).
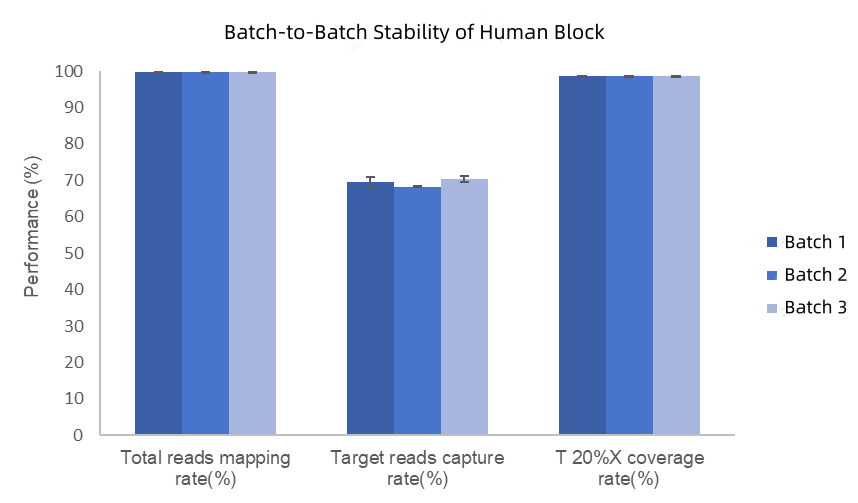
Figure 3. Comparison of blocking effects among different batches of IGT Human Block. The test was carried out using a 170Kb panel and the TargetSeq One v2.0 Capture Kit. A 2 - hour rapid hybridization was performed, and sequencing was conducted on the NovaSeq 6000 platform with PE150.
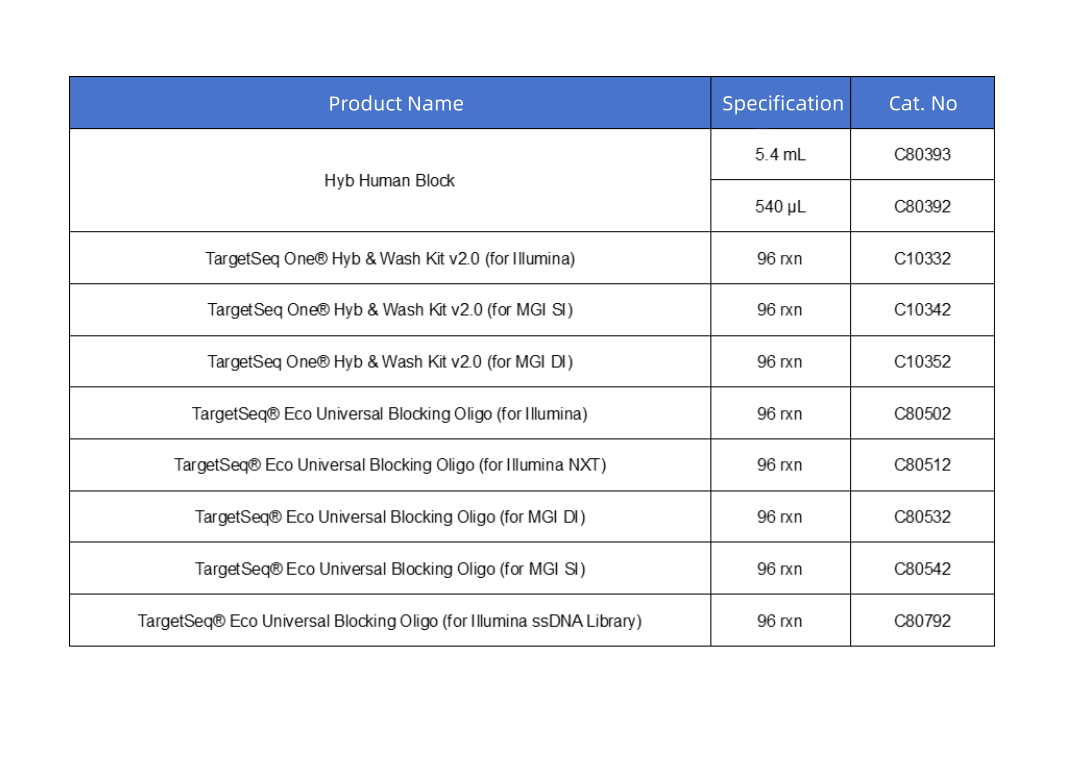
Order Information
Reference
1, Zwick MS, et.al. A rapid procedure for the isolation of C0t-1 DNA from plants. Genome. 1997 Feb;40(1):138-42. doi: 10.1139/g97-020.
2, Duncavage EJ, et.al. Hybrid capture and next-generation sequencing identify viral integration sites from formalin-fixed, paraffin-embedded tissue. J Mol Diagn. 2011 May;13(3):325-33. doi: 10.1016/j.jmoldx.2011.01.006.

 CN
CN





