In recent years, pathogenic microorganisms such as COVID-19, influenza, norovirus, and Mycoplasma pneumoniae have frequently caused clustered infection incidents, which has led to an increasing level of public attention to pathogenic microorganism infections. Relevant health and disease control departments in China have also established an information system for collecting the information on the transmission trends of infectious pathogenic microorganisms. This system enables the continuous dynamic monitoring of pathogens and improves the ability to detect, warn about, and prevent and control sudden infectious diseases.
Figure 1. Some Global Pandemic Incidents of Respiratory Pathogenic Microorganisms
To make timely use of the above-mentioned monitoring system, it is necessary to detect the group pathogens that may emerge at any time in a timely and reliable manner, and accurately determine information such as the source, typing, and evolutionary process of the pathogens. NGS (Next Generation Sequencing, the second-generation sequencing technology) has been widely applied in the fields of pathogen identification, classification, and identification of clinical infectious pathogens due to its advantages such as rapid capture and high sensitivity.
However, the isolation and culture of pathogenic microorganisms face numerous difficulties, and the culture of some highly pathogenic microorganisms poses relatively high risks. Therefore, only some pathogenic bacteria have the conditions for whole-genome sequencing. The rise of targeted capture sequencing technology has provided a new idea for the full-length sequencing of pathogenic bacteria: without going through the culturing process, directly screening, enriching, and sequencing the nucleic acids of pathogenic microorganisms can fundamentally solve the dilemma of culturing pathogenic bacteria, simplify experimental steps, and reduce the risk of infection during experimental operations. Capturing the key sequences of specific pathogens can also accurately identify the types of pathogens in the samples within a short period of time, which greatly speeds up the identification process of pathogens, and is particularly important for quickly responding to the prevalence of infectious diseases.
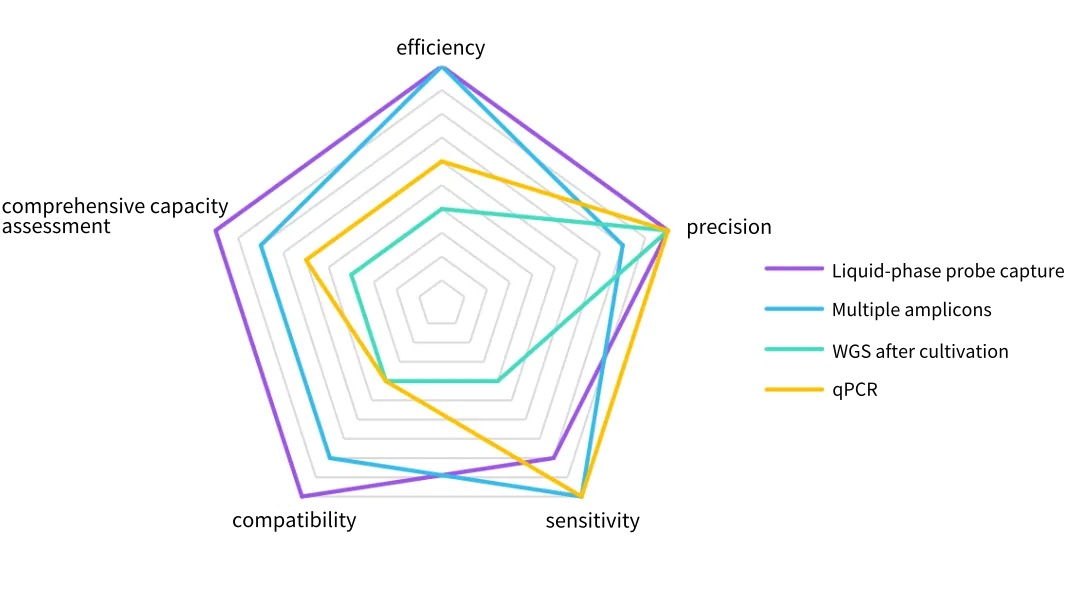
Figure 2. The advantages of different technical platforms in the detection of pathogenic microorganisms and whole-genome sequencing. Compared with traditional cultivation and isolation methods, the NGS capture technology can process a large number of samples in a relatively short period of time. Each sample can obtain extremely high sequence coverage, improving the accuracy of identification and analysis. In addition, the capture technology enhances the ability to detect low-abundance and unknown pathogens, and has a very wide range of applications.
Although the NGS capture technology has great application potential in the field of pathogenic microorganisms, it still faces some technical challenges, such as the design and optimization of probes, the extraction efficiency of pathogen DNA/RNA from complex samples, and the complexity of data analysis. Since its establishment, iGeneTech has been focusing on the field of targeted capture. In response to the urgent need for full-length sequencing of pathogenic microorganisms, it has innovatively proposed a unique design concept for probes of pathogenic microorganisms and developed a number of whole-genome capture products for pathogenic microorganisms.
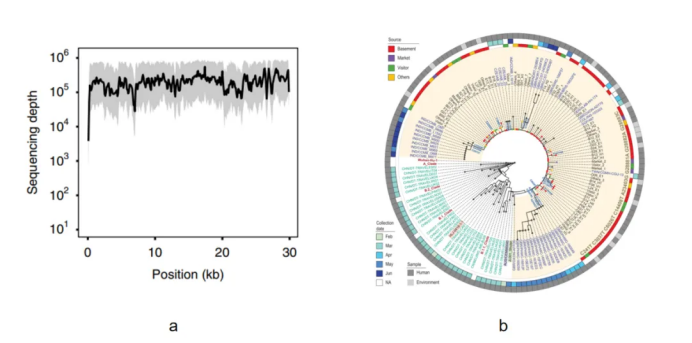
Figure 3. Schematic diagram of the design of iGeneTech's whole-genome capture probes for pathogenic microorganisms. The probes are designed by referring to multiple full-length sequences in the reference database of target pathogenic microorganisms to ensure the sequence diversity of the probes, thereby improving the capture efficiency and coverage of sequences from different strains.
Since 2017, iGeneTech has been continuously developing a variety of whole-genome capture products for pathogenic microorganisms. To date, there are more than 20 predefined panels for pathogenic microorganisms, gradually forming a systematic product matrix covering multiple hosts and systems.
Discovery and traceability of novel pathogenic microorganisms
At the beginning of the COVID-19 pandemic, there was limited sequence information about the novel coronavirus. Based on its profound understanding of the liquid-phase probe capture technology, iGeneTech designed and developed a full-length capture kit for the novel coronavirus by referring to the sequences of closely related strains. This provided technical support for tracing the origin of the sudden epidemic and conducting evolutionary analysis in the early stage. Collaborating institutions also published articles in journals such as Nature Communications and National Science Review at the fastest speed [1,2].
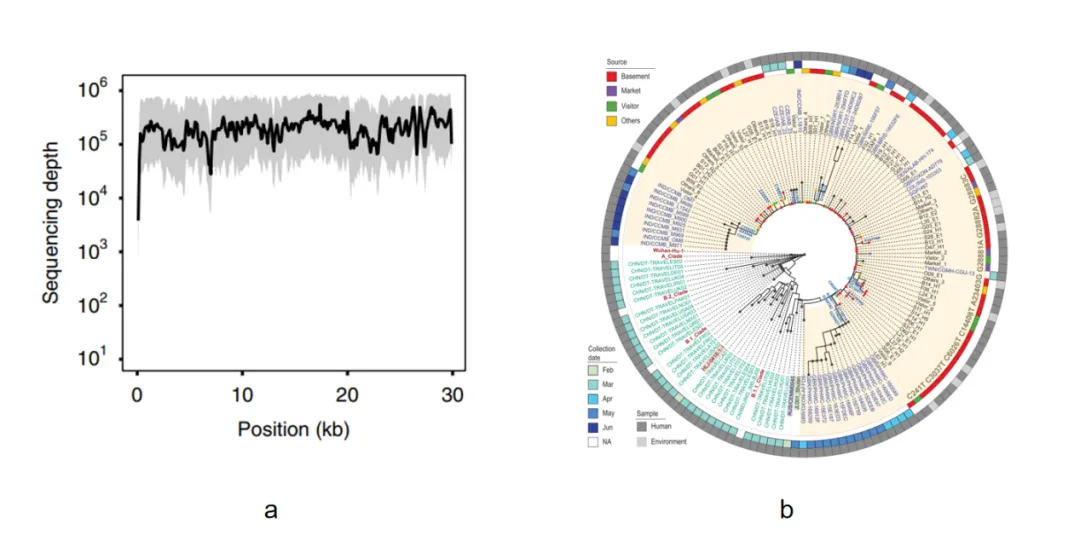
Figure 5. a. The average coverage depth of the full-length capture data of 102 sequenced cases. The shaded area represents the upper and lower quartiles [1]. b. Evolutionary analysis of the full-length SARS-CoV-2 sequences [2].
Daily monitoring of respiratory pathogenic microorganisms
Since 2023, there have been varying degrees of clustered infection events of influenza A, Mycoplasma pneumoniae, norovirus, etc. across the country. The disease control departments have put forward requirements for respiratory disease monitoring. Based on its experience in microbial probe design and a mature hybridization capture system, iGeneTech has custom-developed a number of ultra-fast hybridization capture kits for pathogenic microorganisms, which have been well-received by a large number of customers.
In addition, in response to the diverse daily monitoring needs of disease control departments, iGeneTech has developed a number of predefined whole-genome capture panels related to the respiratory system. Blind tests were conducted on multiple clinical samples with low pathogen loads, and successful typing was achieved and full - length sequences of pathogenic microorganisms were obtained (Figure 6).
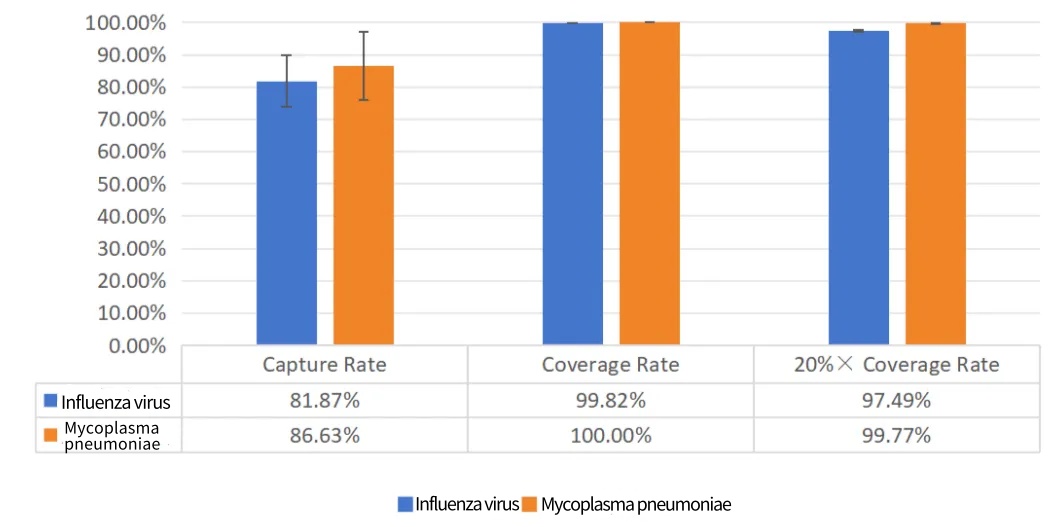
Figure 6. Test data of standard products for whole-genome capture of influenza virus and Mycoplasma pneumoniae.
Capture Rate: Capture efficiency. In the whole-genome capture panel for microorganisms, it is significantly influenced by the load of pathogenic microorganisms in the sample. The lower the load, the lower the capture efficiency usually is. Additionally, it is also affected the size of the microbial genome. The larger the microbial genome and the higher the copy number ratio relative to the host, the higher the capture efficiency.
Coverage Rate: It refers to the coverage of the typed standard sequence by the sample data.
0% Coverage Rate: The coverage rate at 0.2 times the average depth, which is commonly used to evaluate the uniformity of the data. Taking an average depth of 1000× as an example, 20% × Coverage Rate means the 200× coverage rate.
References
1. Du P, Ding N, et al. Genomic surveillance of COVID-19 cases in Beijing. Nat Commun. 2020 Oct 30;11(1):5503. doi: 10.1038/s41467-020-19345-0.
2. Pang X, Ren L, et al. Cold-chain food contamination as the possible origin of COVID-19 resurgence in Beijing. Natl Sci Rev. 2020 Oct 23;7(12):1861-1864. doi: 10.1093/nsr/nwaa264.