
Forensic DNA phenotyping (FDP) inference conducts correlation analysis based on the relationship between specific phenotypes (such as height, skin color, eye color, etc.) and related genetic variations. It uses the genetic information of biological samples to predict the phenotypic characteristics of individuals, so as to assist in solving criminal cases. Human height is a highly heritable polygenic trait. Some studies have shown that the methylation of the P2 promoter of the IGF1 gene affects the concentration of serum growth hormone and human height [1-2]. In addition, both the methylation level and height are continuous variables, and it may be easier to predict height by detecting the DNA methylation status [3].
Hebei Medical University utilized iGeneTech's BisCap® Methylation Capture Sequencing Technology to study the relationship between DNA methylation and human height. In February 2024, the article "The association between DNA methylation and human height and a prospective model of DNA methylation-based height prediction" [1] was published in Human Genetics (Impact Factor: 5.3).

The research team analyzed the DNA methylation array data of 355 individuals of European ancestry. Using the linear regression method, they identified 11,730 differentially methylated sites associated with height. They designed a methylation panel containing 959 CpG sites, with a target region of 123.23 kb. Subsequently, the team recruited 190 participants. They collected whole-blood samples, extracted genomic DNA (gDNA), and used iGeneTech's complete set of methylation liquid-phase hybridization capture system for high-throughput methylation sequencing to construct a height inference model.
The results showed that when using the DNN model to predict height, after applying KPCA analysis to all samples, the root-mean-square error (RMSE), mean absolute error (MAE), and coefficient of determination (R²) in cross-validation were 5.62 cm, 4.45 cm, and 0.64 respectively. The prediction results indicated an obvious sexual dimorphism in the relationship between DNA methylation and height. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses showed that the genes and pathways associated with the selected CpG sites were related to height development. This study demonstrated the association between height and DNA methylation status through epigenome-wide association study (EWAS) analysis. The researchers proposed that targeted methylation sequencing of only 959 CpG sites combined with deep - learning technology can provide a more accurate model for estimating human height than SNP-based prediction models.
Table 1. Prediction errors of all samples.
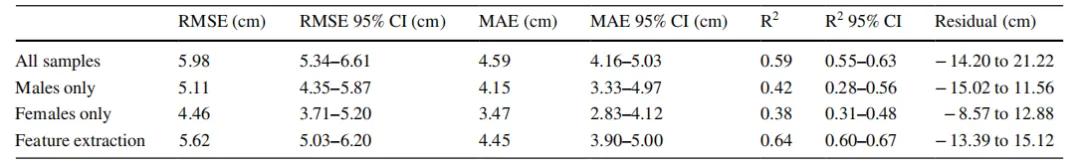
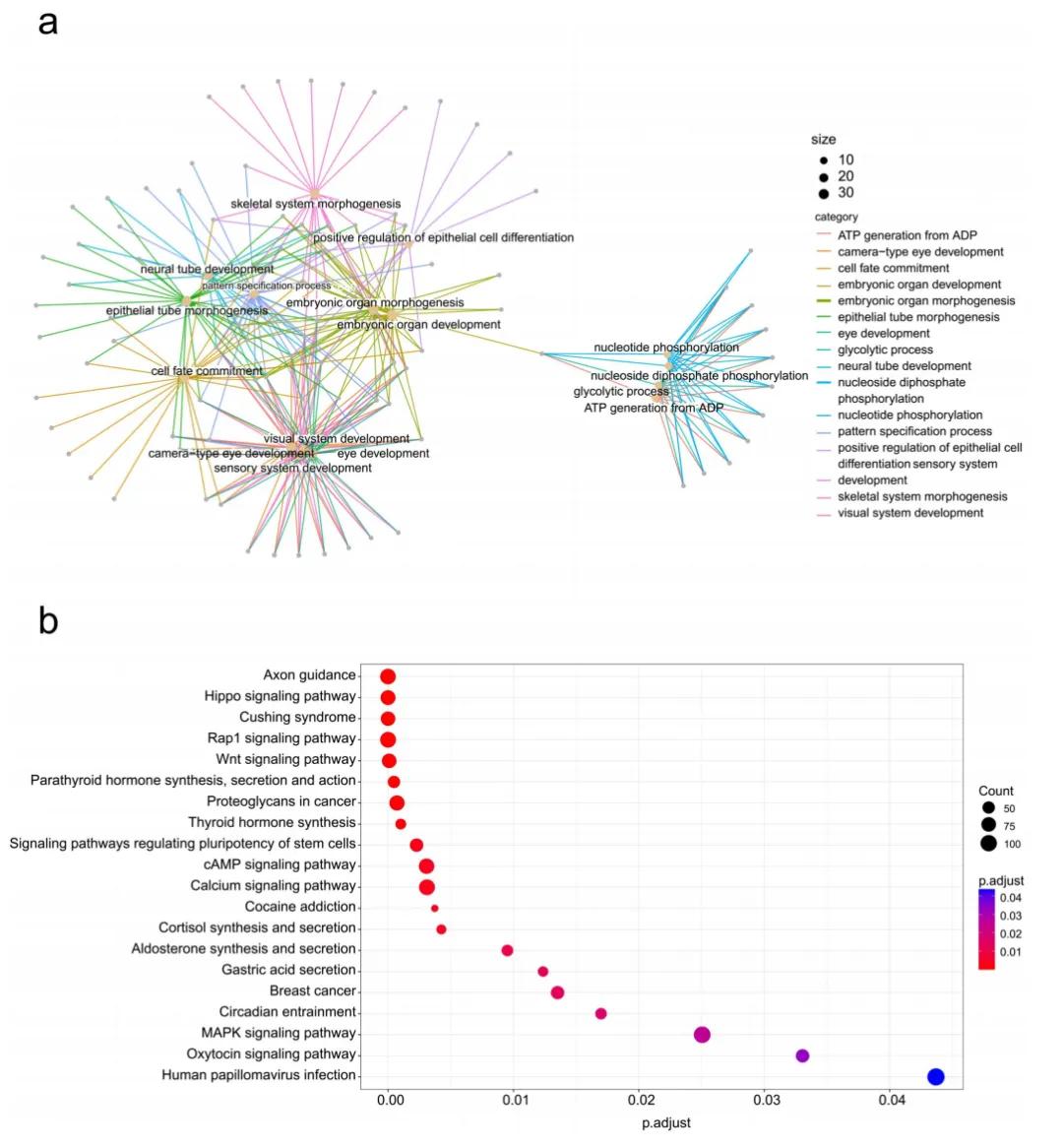
Figure 1. Enrichment analysis of GO functions and KEGG pathways.
BisCap® Methylation Capture Sequencing Technology
Based on the research and development of TargetSeq® hybridization capture sequencing technology, iGeneTech has perfectly combined the target region capture technology with methylation sequencing, achieving full coverage of methylation sites in the target region. It can be applied to different sample types such as gDNA, FFPE DNA, and cfDNA. Meanwhile, while taking into account the detection cost, it provides higher sequencing depth, improving the sensitivity and accuracy of detection.
01 Probe Design
The panel design for CG sites on both the positive and negative strands can reduce costs and ensure the methylation detection rate. iGeneTech has launched a variety of methylation products, including a new solution for the research and development of tumor methylation early screening markers-BisCap® Human CpG Island Panel, and a multi-cancer methylation early screening and detection product-BisCap® Multi-Cancer Early Detection Panel. When the methylation products are used in combination with iGeneTech's self-developed single-strand library preparation kit, they can efficiently capture trace methylation signals in the blood, providing more support for clinical early screening and detection of tumors.
02 Library Preparation Scheme
IGT® ssDNA Library Prep Kit is a universal single-strand DNA (ssDNA) library preparation kit. This product can be used for the construction of methylation libraries starting from 1 ng to 50 ng of DNA. During the construction of the methylation library, bisulfite treatment will cause serious DNA damage, including fragmentation, degradation, and unwinding, etc. In addition, there is a large amount of single-strand DNA in low-quality or severely degraded DNA. This kit can perform adapter ligation on ssDNA and is suitable for the screening of methylation markers, early tumor screening, and other aspects.
It is suitable for low-input samples such as cfDNA and FFPE DNA, and the starting amount for library preparation can be as low as 1 ng.
It has a high cfDNA recovery rate, a double-ended Poly structure, and is compatible with DNA damage.
The highly optimized experimental process can effectively improve the complexity of the library and the validity of the data.
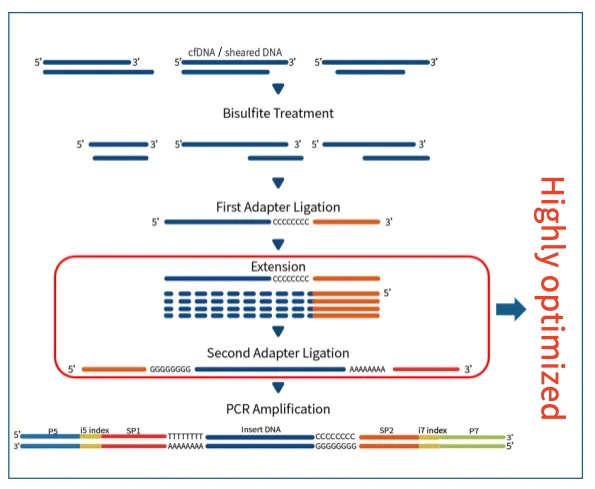
Figure 2. Flow chart of the methylation library preparation process using IGT® ssDNA Library Prep Kit.
IGT® ssDNA Library Prep Kit has a high cfDNA recovery rate. Through comparative tests with other similar kits on the market, experiments were carried out using 5 ng cfDNA samples. The results showed that this kit can recover these small fragment cfDNAs more effectively, thus providing richer data in methylation analysis and related research.
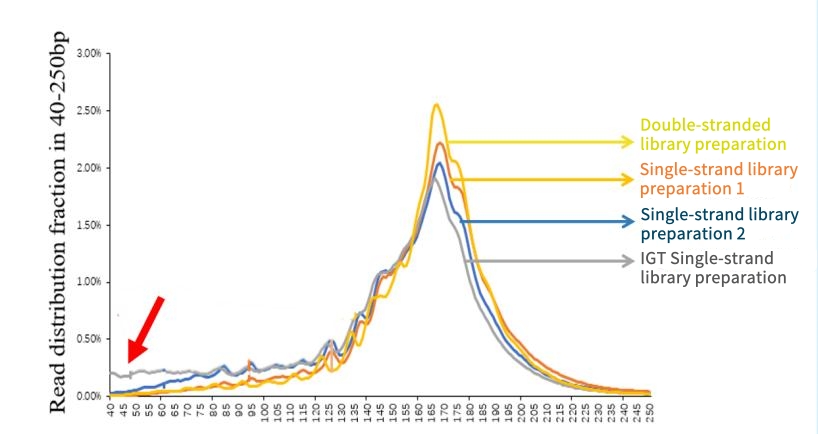
Figure 3. The recovery rate of cfDNA samples by different methylation library preparation kits.
03 Methylation Hybridization Capture System
The self-developed BisCap® Enhancer can effectively improve the capture efficiency. Due to the unknown methylation status of different CpG sites, abnormal GC content, and other reasons, there is a situation of low capture efficiency in the methylation library. Adding BisCap® Enhancer to the BisCap® methylation hybridization system can increase the capture efficiency by 10%.
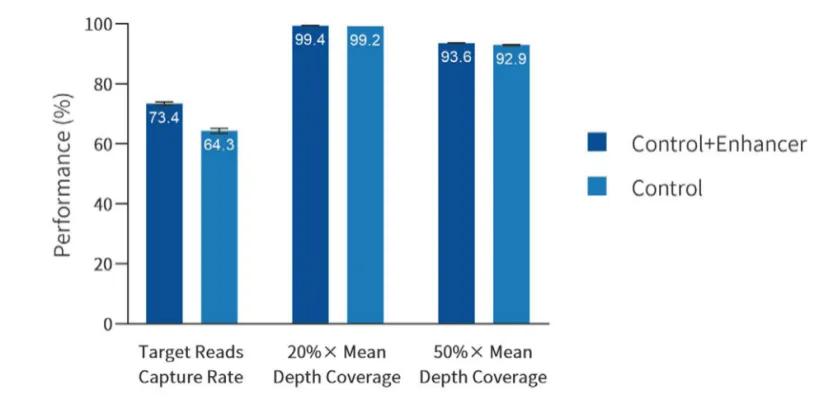
Figure 4. The data performance of adding BisCap® Enhancer to the BisCap® hybridization elution system. The sample is the DNA of the NA12878 cell line. 200 ng of the sample is used for library preparation. The BisCap® Fast Library Prep Kit and IGT® Methyl Adapter & UDI Primer (for Illumina) are applied for library construction. A 123 kb panel is used for capture, and sequencing is conducted on the NovaSeq 6000 platform with PE150 mode.
References
1.Ouni M, Belot MP, Castell AL, Fradin D, Bougneres P (2016a) The P2 promoter of the IGF1 gene is a major epigenetic locus for GH responsiveness. Pharmacogenomics J 16:102–106.
2.Ouni M, Castell AL, Rothenbuhler A, Linglart A, Bougneres P (2016b) Higher methylation of the IGF1 P2 promoter is associated with idiopathic short stature. Clin Endocrinol 84:216–221.
3.Wang Z, Fu G, Ma G, Wang C, et al . The association between DNA methylation and human height and a prospective model of DNA methylation-based height prediction. Hum Genet. 2024 Mar;143(3):401-421.