Index (also known as Barcode) plays a critical role in high-throughput sequencing as a unique nucleotide sequence used to distinguish different samples. It is added to the sequencing libraries of DNA or RNA fragments from each sample to facilitate data demultiplexing and sample identification of the sequencing data. With the rapid development of next-generation sequencing (NGS) technology, its high-throughput nature enables massive data generation in a single run, leading to an increase in the number of samples processed per sequencing run. To effectively distinguish and identify the sequencing data of each sample, a greater variety of Index types is required.

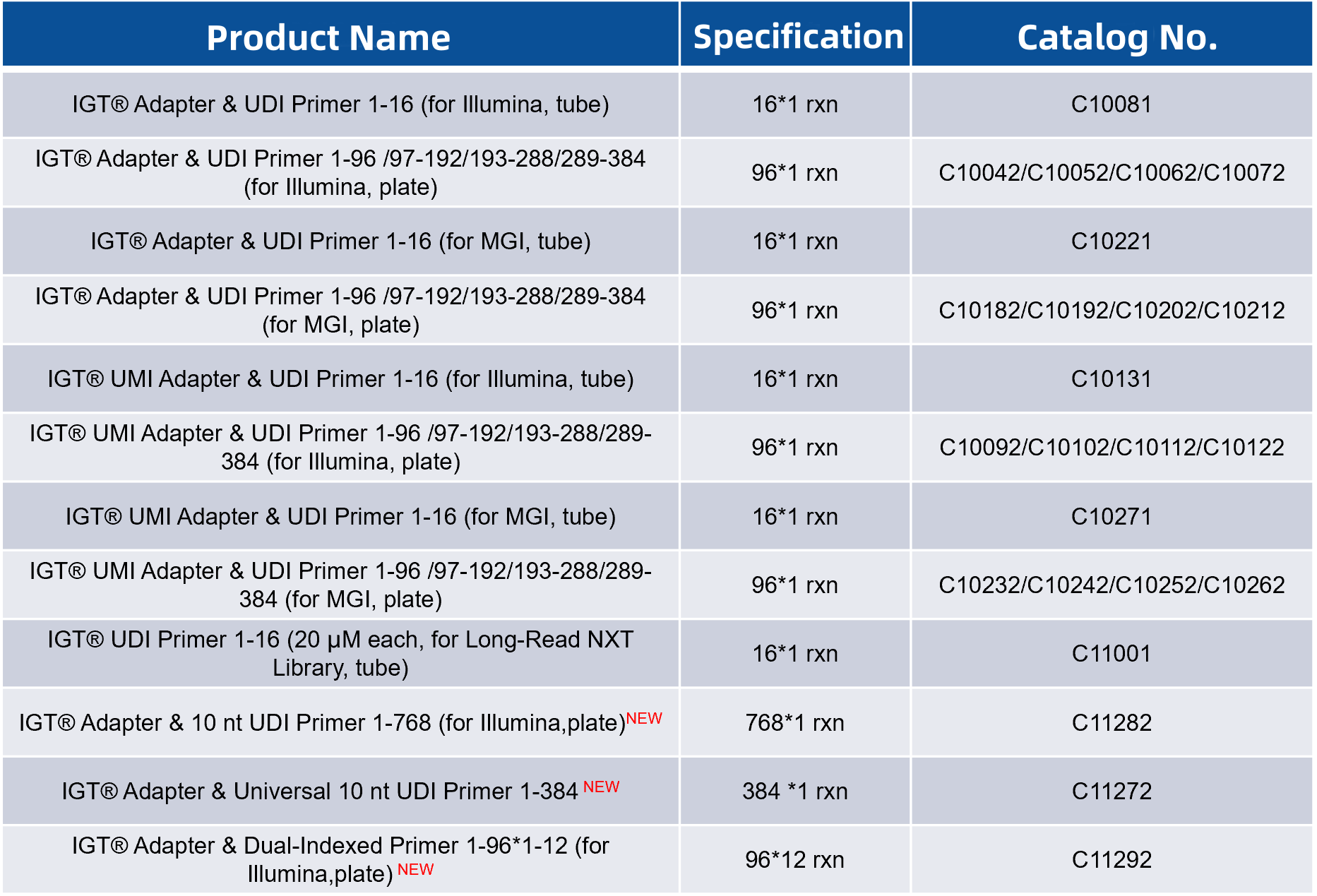
Table 1. Full names and abbreviations of common Index types
Since 2020, iGeneTech has been continuously adding new "members" to the Index product series. Currently, it has developed product series including up to 768 types of UDI and 4,608 types of CDI. These products are compatible with various adapter library preparation solutions to meet the requirements of high - throughput sequencing with a large number of samples in a single run.

Figure 1. Summary of iGeneTech Index Product Series
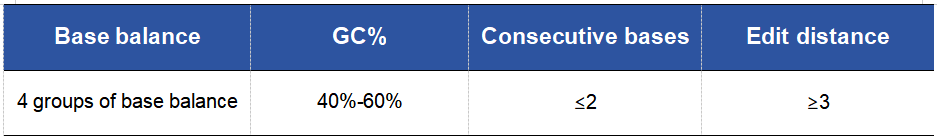
Table 2. 10nt UDI Design Principles
Note:
· Base balance: It refers to the balance degree of Index. The proportions of the four bases A, T, C, and G in a set of Index need to be balanced to improve the accuracy of sequencing and reduce signal deviation during the sequencing process.
· GC content: The GC content of Index should be maintained between 40% and 60% to ensure good amplification efficiency and sequencing quality during PCR amplification and sequencing.
· Edit distance: Even when one base is substituted, deleted, or inserted in the Index sequence, an edit distance of 3 must be strictly maintained. A larger edit distance can ensure sufficient distinguishability between them, thereby reducing mis-attribution during the sequencing process.
The 768 types of UDI enable each sample to have a unique "cap" and a unique "shoe" even when conducting large - scale sample testing. Samples can be distinguished by their matching "caps" and "shoes", which helps to more accurately disassemble the corresponding relationship between sequencing data and samples and reduces the probability of samples being "mislabeled".
1. Uniform and stable library yield: Under the same experimental conditions, the average library yields are basically the same. Each UDI has an extremely high and uniform independent amplification efficiency.
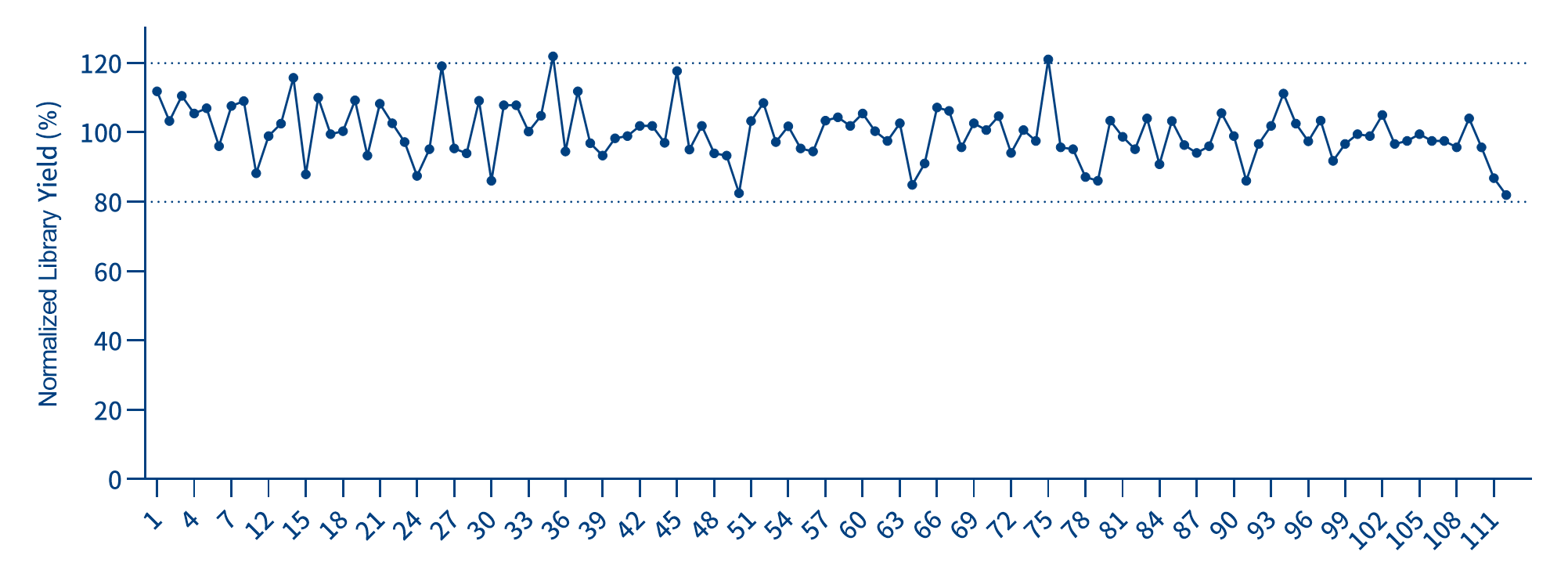
Figure 2. Demonstration of Library Yield
*Randomly select a set of indexes from IGT® Adapter & 10 nt UDI Primer 1 - 768 (for Illumina, plate). Add 50 ng of gDNA standard to each reaction, and use the IGT® Fast Library Prep Kit V2 to construct WGS libraries. The output of the libraries constructed from each sample is stable and uniform.
2. Uniform and effective demultiplexing rates and data output
When multiple samples are pooled and sequenced in a single lane, each UDI maintains a consistently high effective demultiplexing rate, ensuring uniform data output across samples.
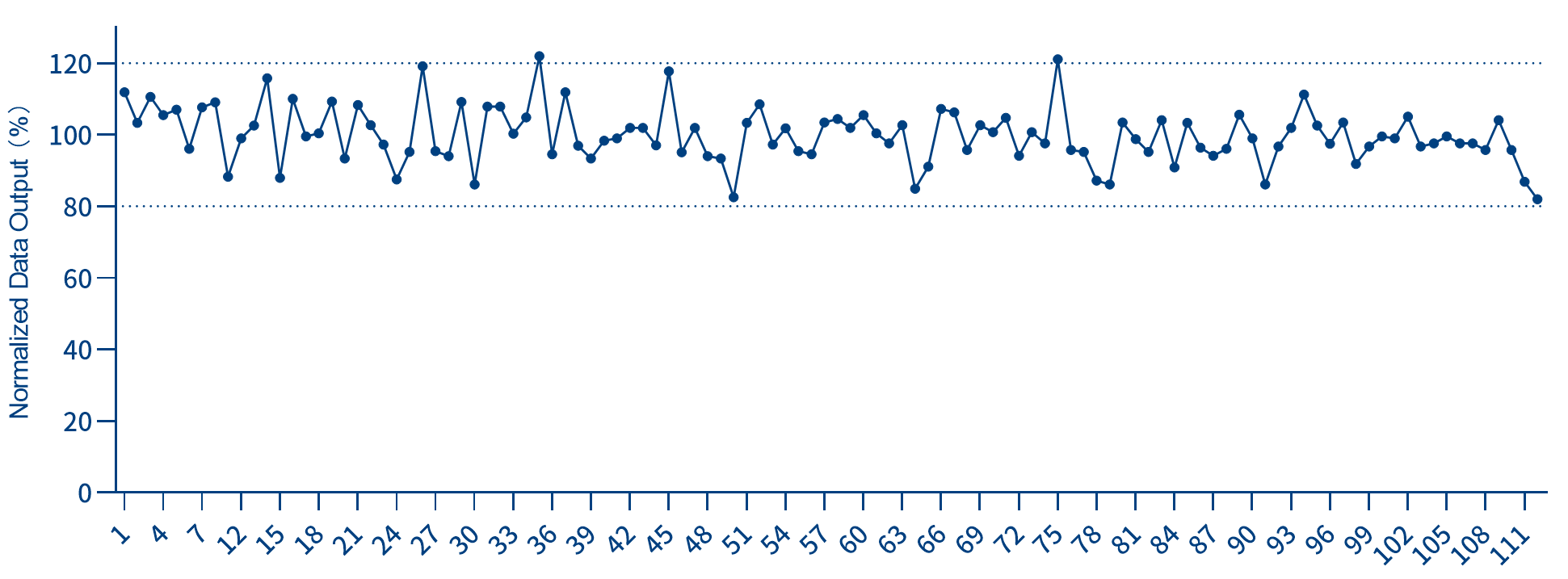
Figure 3. Demonstration of the effective demultiplexing rate of UDI
*Randomly select a set of Indexes from IGT® Adapter & 10 nt UDI Primer 1 - 768 (for Illumina, plate). Add 50 ng of gDNA standard to each reaction, use iGeneTech Fast Library Prep Kit V2 to construct WGS libraries, and conduct sequencing on NovaSeq 6000 with PE150.
The more flexible plate-based packaging for I7 and I5 Indexes is compatible with automated equipment for mixed use, enabling up to 4,608 Index combinations and facilitating convenient, accurate, and efficient loading of ultra-large sample quantities.