With the continuous development of scientific research and the needs of precision medicine, large-scale gene information detection based on next-generation sequencing (NGS) technology has become an important research method. The accuracy and traceability of data have become the top priority in large-scale data collection and analysis. However, confusion and contamination can easily occur in sample collection, transportation, splitting, and experiments, leading to gene information errors and causing enormous losses to research and clinical applications. Investigations reveal that nearly 80% of researchers have encountered problems with sample confusion, and other researchers analyzing some publicly published human gene data found that about 3% of samples were confused.
To more quickly discover and track such sample issues, many guidelines and expert consensuses recommend that testing institutions set up sample tracking processes and perform mandatory consistency analysis on data and samples. Only samples that pass these checks can be uploaded for subsequent bioinformatics and genetic information analysis. For example, the expert consensus on the application of whole exome sequencing technology in prenatal diagnosis (see the image below) explicitly describes the requirements for sample consistency.

Similarly, the College of American Pathologists (CAP) has specific regulations on the possibility of cross-contamination in their sample handling and aliquoting requirements for molecular diagnostic laboratories:
MOL32360: Specimen Handling: There are written procedures to prevent specimen loss, alteration, or contamination.
MOL32385: Specimen Aliquots: If aliquoting of specimens is performed, there is a written procedure to prevent any possible cross-contamination of the specimens.
In practice, sample tracking and quality assurance mainly include two aspects: 1) Gender consistency identification; 2) Single nucleotide polymorphism (SNP) consistency identification: It is recommended to select multiple SNP loci or other tags as sample identification (sample ID).
Based on two independently owned technology platforms, TargetSeq hybrid capture sequencing technology and MultipSeq multiple amplicon sequencing technology, iGeneTech has designed and developed liquid phase probes for 100 high MAF (Minor Allele Frequency) SNP loci, which can be added to customized liquid phase probes. At the same time, iGeneTech has developed a multiple amplicon quality control validation kit for these 100 loci: MultipSeq Sample ID Research Assay. By consistently comparing the genotyping results from both multiple amplicon and liquid phase methods for the 100 SNPs, each sample can be explicitly tracked throughout the detection process, monitoring contamination between samples and avoiding human operational errors.
This panel is suitable for various sample types and low starting amount samples, supporting direct blood amplification, providing an accurate, simple, and economical solution for the confusion and contamination issues within the laboratory quality management system.
iGeneTech offers the option to add the 100 SNP quality control probes to custom liquid capture products for free. Additionally, by purchasing the MultipSeq Sample ID Research Assay, sample quality control verification can synchronize with accurate sample identification. According to the likelihood ratio formula calculation and evaluation in the Ministry of Justice's "Individual Identification Technical Specification--SFZ JD0105012--2018," 100 SNPs are sufficient to achieve identification within the Chinese population.
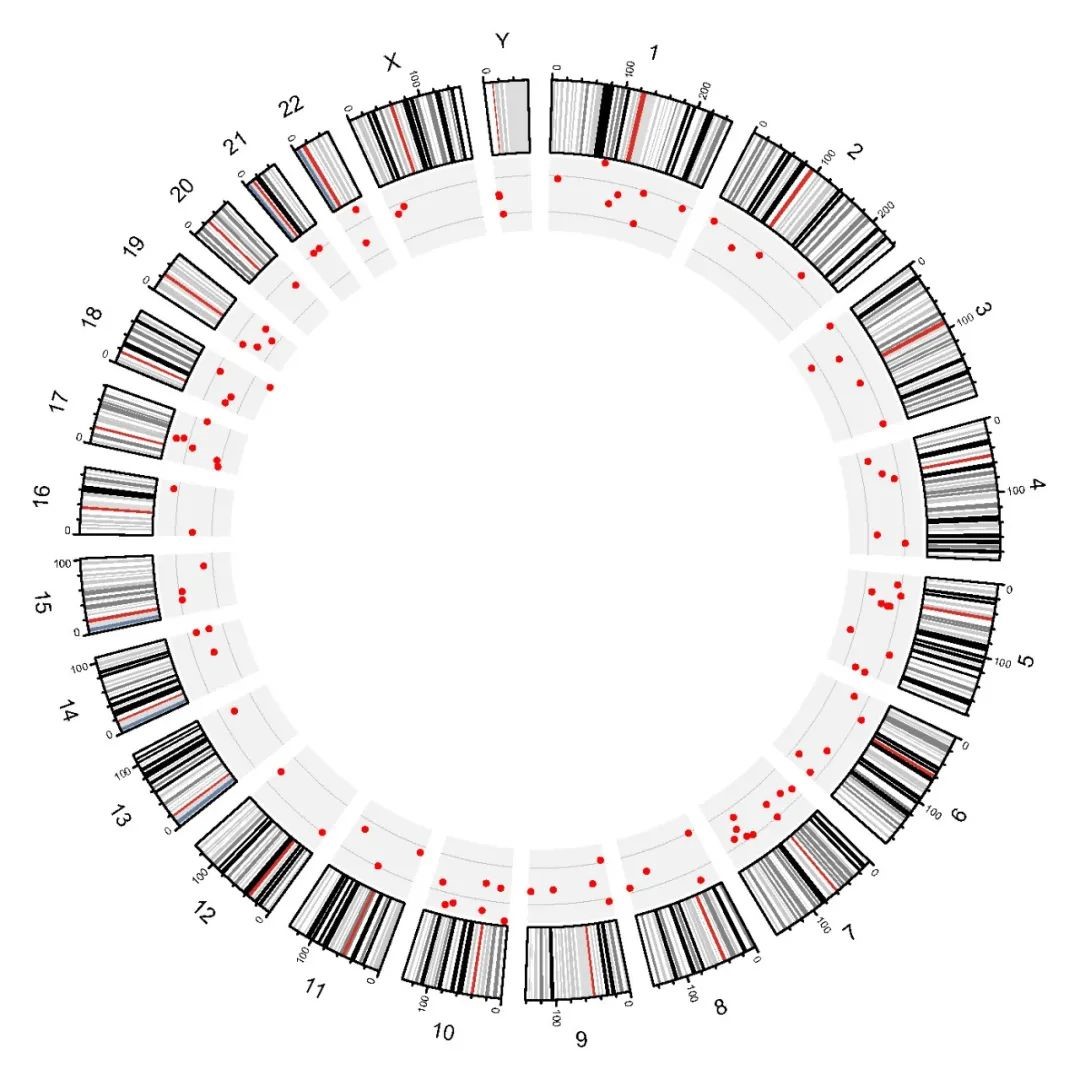
Figure 1. Genomic distribution of the 100 SNP quality control loci
Individual identification does not rely on a single genetic marker but rather the combined use of a set of independently inherited genetic markers to identify different individuals within a population, known as total discrimination power (TDP). The closer the TDP value is to 1, the stronger the individual identification ability of the set of genetic markers.
The cumulative individual identification power calculation formula:
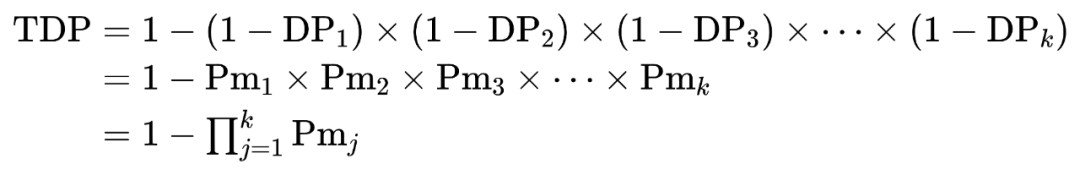
k is the number of genetic markers, Pmj is the Pm value of the j-th genetic marker in the detection system.
where DP is the individual identification power of a single genetic marker, calculated as:
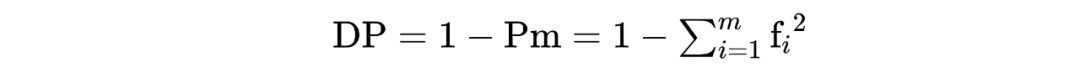
m is the frequency of a certain genetic marker's phenotype, fi is the frequency of the i-th phenotype.
For example, selecting high MAF SNP loci as the 100 quality control loci, at an MAF of 0.5, the higher the DP and TDP values, the stronger the individual identification power, and the better the quality control ability for sample confusion and contamination.
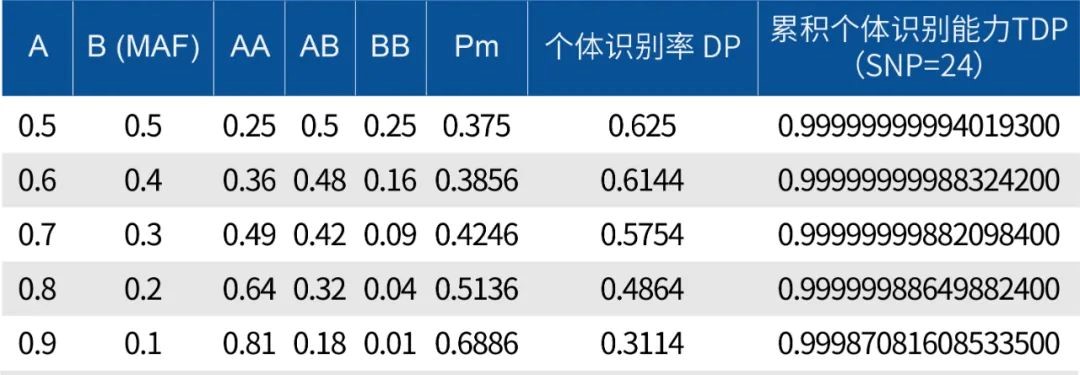
According to the likelihood ratio (LR) evaluation in the specification, the larger the LR value, the more it supports the hypothesis that the test sample comes from a certain individual. When the LR numerically exceeds the total global population, the evidence is strong enough to support this result.
At an MAF of 0.5, with 45 SNP loci, LR=1.53E+09, exceeding the total population of China. Therefore, 100 high MAF loci are sufficient for individual identification within the Chinese population, providing higher accuracy for sample tracking.
Product Features
Used for sample identification and tracking research.
Identification capability – able to achieve individual identification within the Chinese population.
Adding the same loci probes to liquid panels allows concurrent precise sample identification
Amplicon sizes of 110~130 bp are suitable for various sample types.
Supports direct blood amplification, suitable for low starting amount samples.
A single tube primer, simple multiple amplicon operation process, compatible with iGeneTech automated equipment.
All exome series products include the 100 loci quality control probes.
Provides an overall solution, including experiment process and data analysis process setup
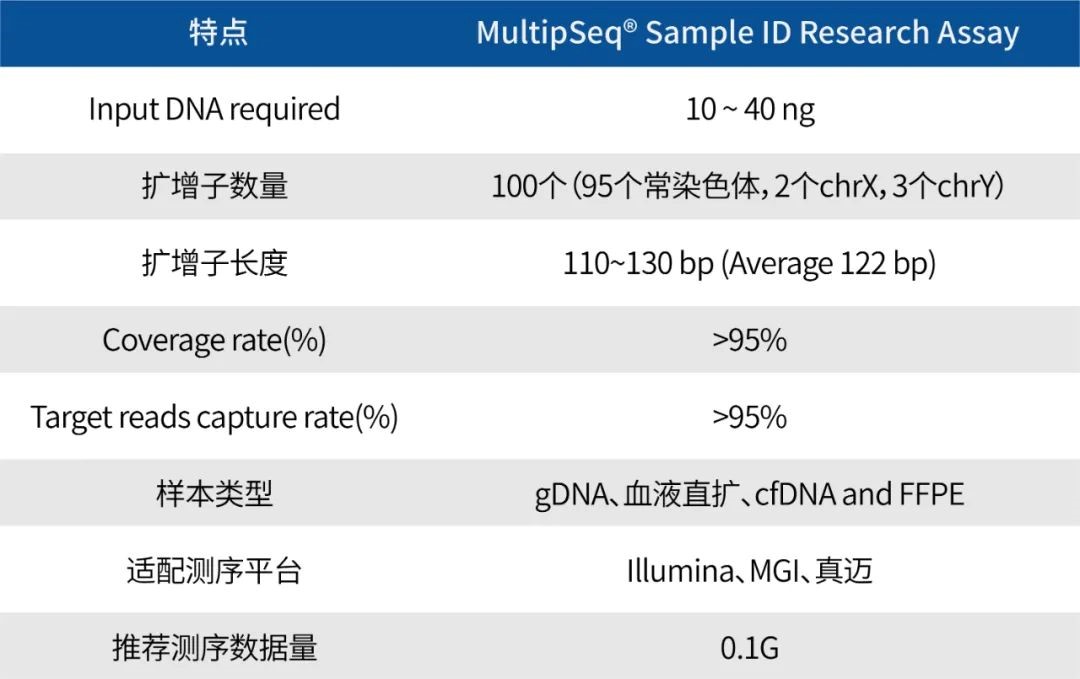
Technical Parameters
Operation Process
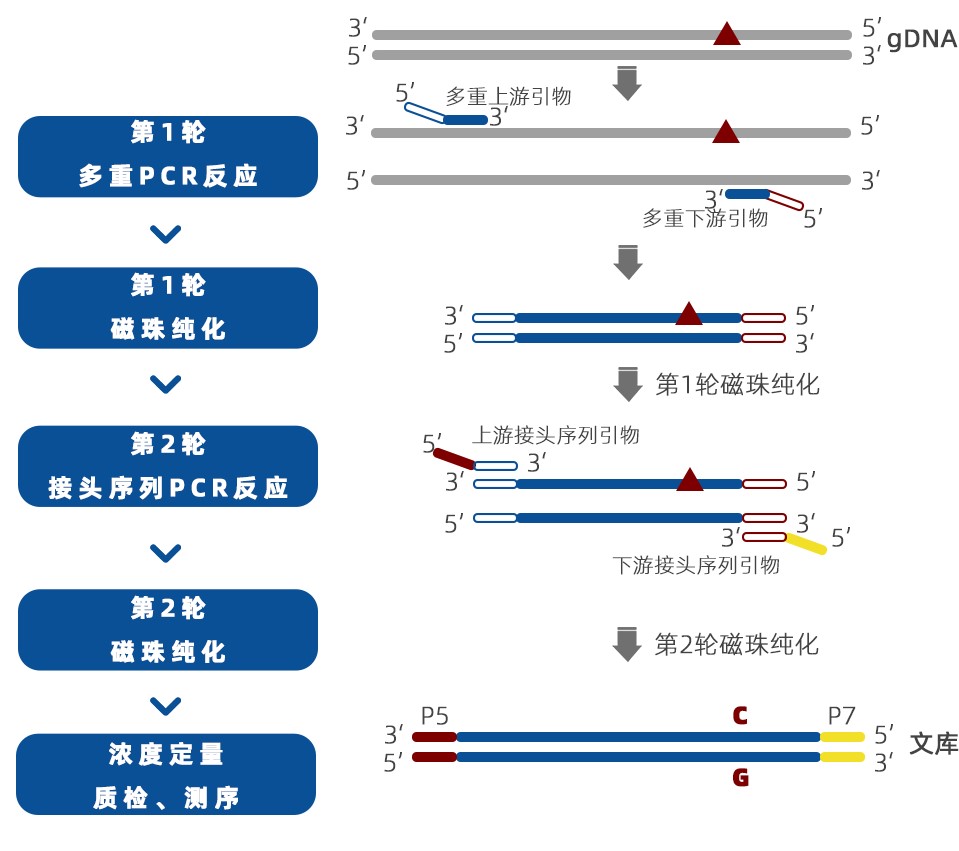
Figure 2. MultipSeq Multiple Amplicon Sequencing Technology Process
This panel adopts the principle of MultipSeq multiple amplicon sequencing technology, performing PCR amplification on the above 100 loci. Through the above process, an NGS sequencing library is obtained, sequenced after quality inspection. The data is then analyzed for genotyping consistency with the same 100 loci in the liquid phase, tracking the possibility of sample confusion and contamination, ensuring sample quality control.
Data Performance
Panel Data Indicators
Using Promaga mixed blood gDNA standard G304A, a male saliva sample, and a female saliva sample, tested with a 40ng input amount. G304A and the male saliva covered 100 loci T 30X coverage rate of 100%, and the female saliva covered all but the 3 Y chromosome loci, covering 97 loci fully.
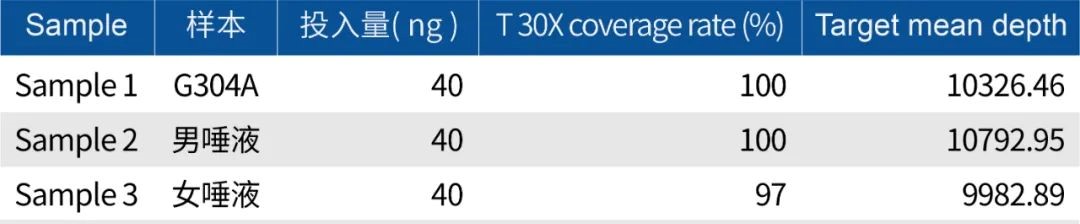
In the actual samples, the coverage rate for male samples was 100%, with excellent capture efficiency and uniformity indicators.
This panel contains 3 loci identification loci on chrY, and the 20X coverage rate for female samples was 97%.
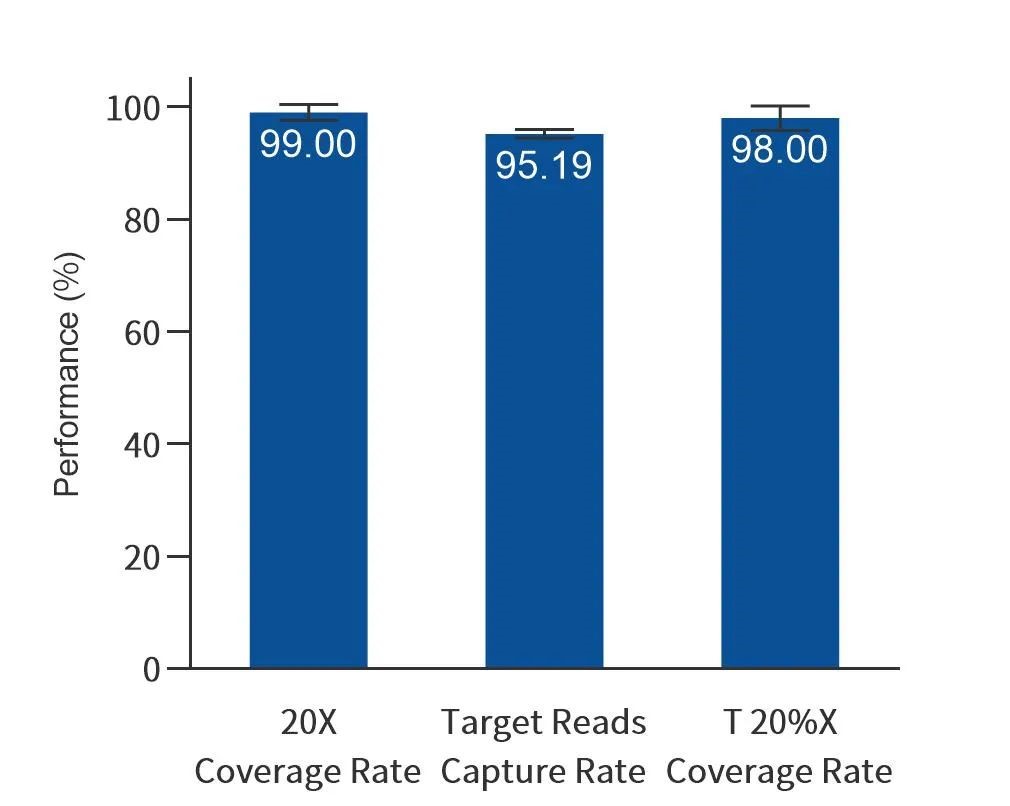
Figure 3. Data indicators for quality control loci in actual samples
High Consistency Between Multiple and Liquid Phase Genotyping
Actual samples were tested for whole exomes with AIExomeV2Plus, and sample quality control using the MultipSeq Sample ID Research Assay. The genotyping consistency for the 100 common loci tested was 100%*.
In female samples, due to the inclusion of 3 loci on chrY, the genotyping consistency was 97%.
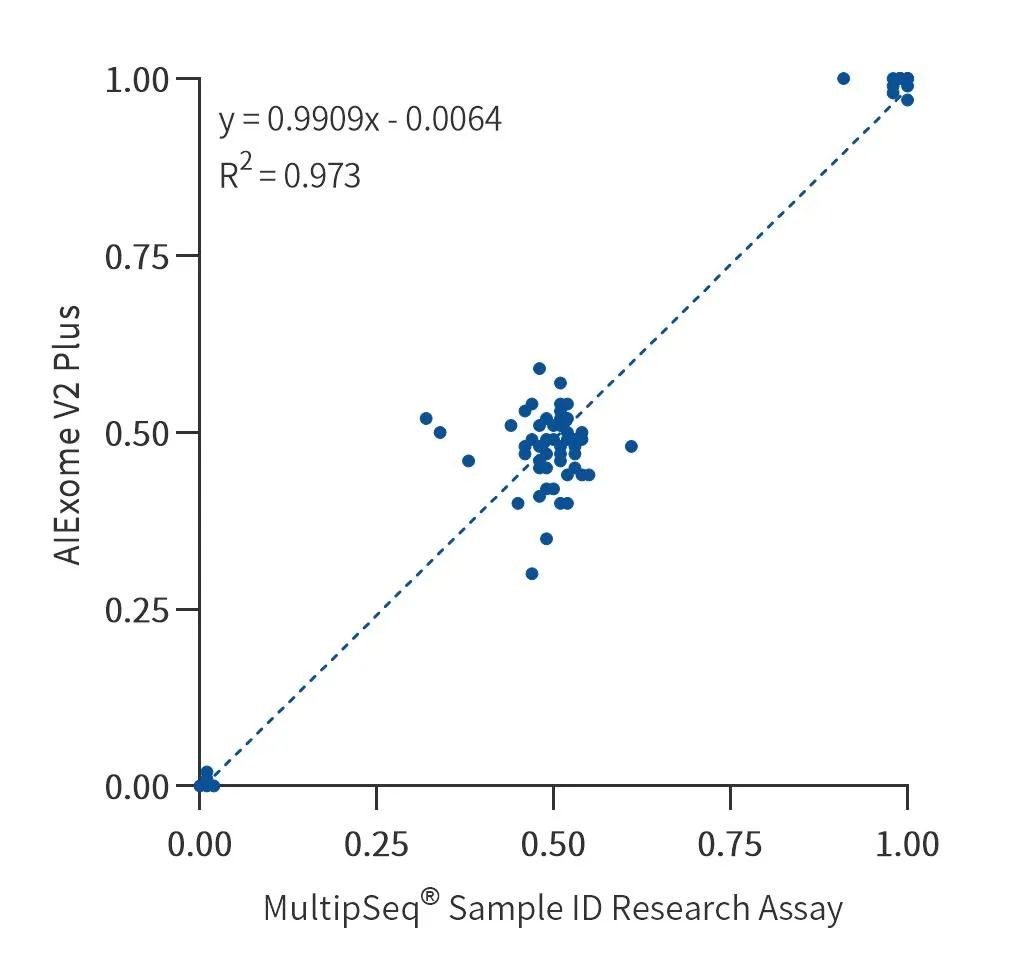
Figure 4. AIExome V2 Plus and MultipSeq Sample ID Research Assay genotyping consistency results for 100 quality control loci in actual samples
Data Performance
Data performance for different sizes of liquid custom panels spiked with 100 quality control probes. Tests were done with liquid custom panels of 258 k, 518 k, and 857 k, with similar capture efficiency.
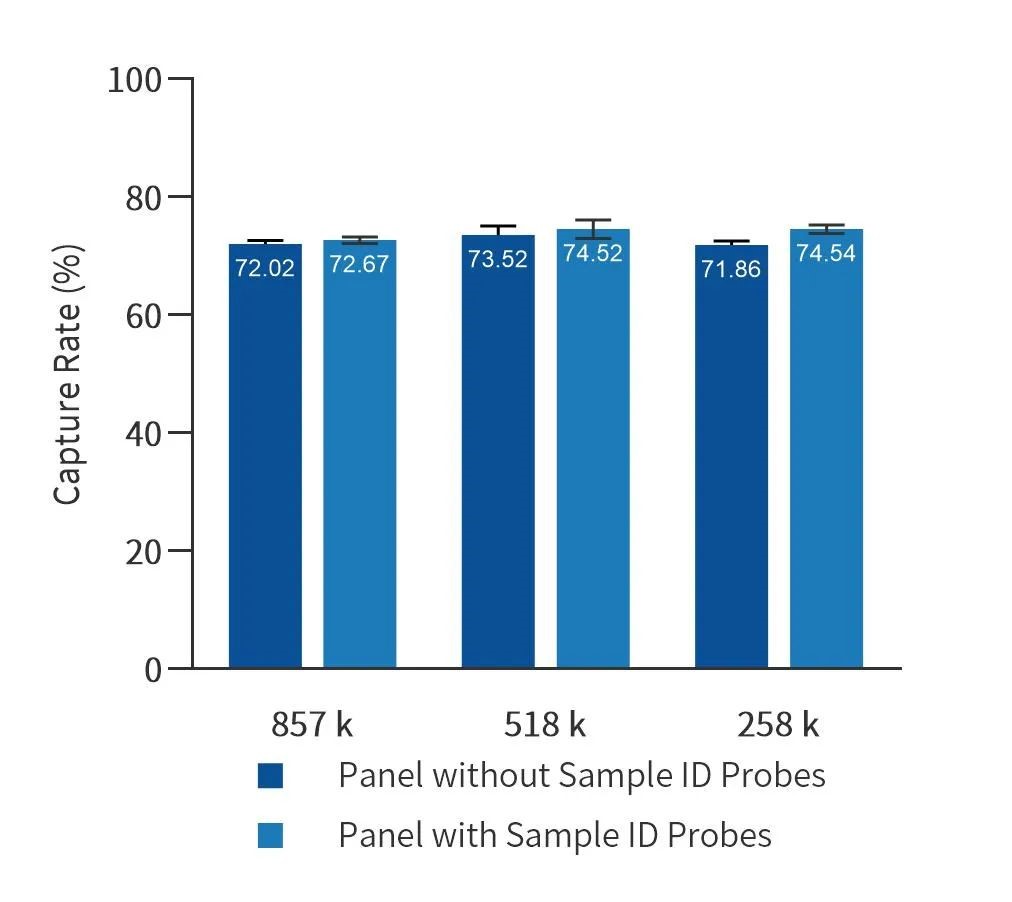
Figure 5. Data performance for 100 quality control probes spiked into different liquid custom panels (258 k, 518 k, and 857 k)
MultipSeq Sample ID Research Assay supports direct blood amplification and low starting amount library construction, compatible with iGeneTech's automated equipment, providing enterprise clients with efficient and convenient sample tracking solutions.
Ordering Information

About iGeneTech
iGeneTech is a national high-tech enterprise focusing on gene capture technology. It owns independently developed gene capture technology platforms for probe hybridization and multiple PCR, and also has the diagnostic service capability for large sample volumes utilizing gene capture technology, committed to providing globally leading overall solutions for gene capture technology.
After years of industrial development in gene capture technology, iGeneTech's product line now fully covers the upstream, midstream, and downstream of the gene capture field, including custom development of personalized gene capture products, gene capture laboratory LIMS system, full-process laboratory automation solutions, data management and interpretation solutions, laboratory quality management system solutions, NGS sequencing ultimate cycle service solutions, and ultra-high-throughput sample diagnostic service solutions. iGeneTech has become a world-leading supplier of comprehensive gene sequencing technology solutions, providing all-cycle, highly automated, intelligent, and productized gene testing ALL-IN-ONE solutions for third-party clinical testing institutions, hospital precision medicine centers, gene testing companies, researchers, and clinicians.