Overview
iGeneTech Bioscience offers 2 full length SARS-CoV-2 genome capture solutions for COVID-19 surveillance and research by using hybridization capture and multiplex amplicon technologies.
iGeneTech Bioscience offers 2 full length SARS-CoV-2 genome capture solutions for COVID-19 surveillance and research by using hybridization capture and multiplex amplicon technologies.
Panel Number | T1302XV1(Hybridization Capture) A186XV6/V7 (Multiplex Amplicon)" |
Technical Platform | T1302XV1: TargetSeq® Hybridization Capture Sequencing
A186XV6/V7: MultipSeq® Multiplex Amplicon Sequencing |
Coverage Size | T1302XV1:510.1 Mb
A186XV6/V7:29.8 kb |
Reference Database | NCBI/Taxonomy |
Reference Genome | 10,000 full-length sequences of SARS-CoV-2 genome |
Coverage | SARS-CoV-2 genome full length |
Storage | T1302XV1: -20℃±5℃ A186XV6/V7:-20℃±5℃ |
Sequence Platform | T1302XV1:Illumina / MGI
A186XV6:Illumina
A186XV7:MGI |
Recommend Sequencing Read Length | PE150 |
Recommend Sequencing Data Size and Depth | T1302XV1:2 Gb
A186XV6/V7:0.2 Gb |
99.9% coverage of SARS-CoV-2 genome for a comprehensive understanding of the virus.
It is suitable for a wide range of samples, such as nasopharyngeal swabs, deep sputum, alveolar lavage fluid, lung tissue biopsy specimens, etc.
High sensitivity MultipSeq® multiplex amplicon technology allows detection limits as low as 50 copies.
Updating panel according to the latest database for known and novel strains.
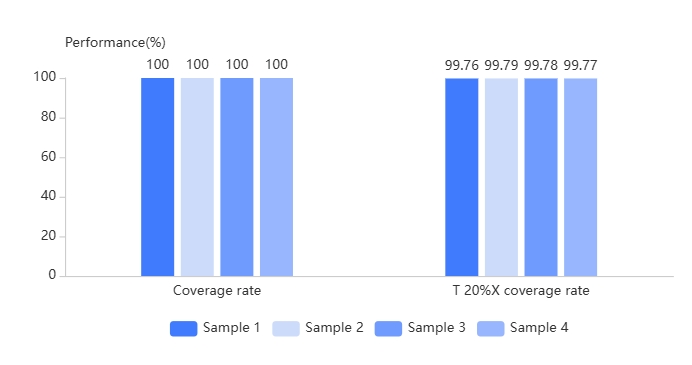
With the proven TargetSeq One® workflow, test results showed high coverage and uniformity for TargetSeq® option.
Test results of MultipSeq® multiplex amplicon option with COVID-19 full length standard sample (Gene-Well, cat.TEST09) showed extremely high detection sensitivity.
| Sample | Input (Copies) | Raw Data (Mb) | Data Efficiency (%) | Mapping Rate (%) | Capture Rate (%) | 1X Coverage Rate (%) | Average Sequencing Depth | 100X Coverage Rate (%) | 20%X Coverage Rate (%) |
| Sample5 | 22000 | 77.88 | 95.49 | 96.94 | 100 | 100 | 2461.28 | 99.23 | 93.31 |
| Sample6 | 2200 | 169.35 | 95.54 | 95.53 | 100 | 100 | 5325.76 | 99.21 | 96.66 |
| Sample7 | 220 | 160.6 | 75.46 | 66.18 | 100 | 100 | 3471.37 | 99.35 | 96.68 |
| Sample8 | 22 | 212.26 | 46.43 | 22.3 | 100 | 100 | 1521.07 | 98.21 | 79.77 |
| Sample9 | 2 | 218.76 | 37.41 | 8.66 | 100 | 98.99 | 606.28 | 92.59 | 80.63 |

Analysis of the genome of severe acute respiratory syndrome coronavirus type 2 (SARS-CoV-2) from clinical samples is critical to understanding virus transmission and evolution and vaccine development. Existing RNA sequencing methods are technically and time-demanding for users and are not applicable to time-sensitive clinical samples, and not optimized for the high performance of the viral genome. In this study, full-length capture of coronavirus was performed after library construction of clinical samples by capture technology, and a complete high-depth, high-coverage SARS-CoV-2 genome with high yield and stability was successfully obtained. By reducing the hands-on time from sample to viral enrichment sequencing library, this rapid, versatile, and clinically friendly method will facilitate molecular epidemiological studies during current and future outbreaks.

The spread of SARS-CoV-2 in Beijing by May 2020 was caused by domestic and global transmission of imported cases. The study reported genomic surveillance data from 102 imported cases, which indicated that all cases in Beijing could be broadly classified into three categories: Wuhan exposure, local transmission, and overseas importation. The study classified all sequenced genomes into seven clusters based on representative high-frequency single nucleotide polymorphisms (SNPs). Genomic comparisons showed that the input group had higher genomic diversity than the Wuhan-exposed and locally transmitted groups, suggesting continued genomic evolution during global transmission. The input group had region-specific SNPs, whereas single nucleotide variants within the host showed a random character with no significant differences between groups. The epidemiological data suggest that case detection in a mandatory quarantine entry situation may be an effective method to prevent the recurrence of outbreaks triggered by imported cases. Notably, the study also identified a novel set of insertions/deletions (InDel). The data from this study suggest that the SARS-CoV-2 genome may have high mutation tolerance.
| Product Name | Panel Number | Set | Cat. No |
| TargetSeq® SARS-Cov-2 Panel | T1302XV1 | 16 rxn | PH2001721 |
| 96 rxn | PH2001722 | ||
| MultipSeq® SARS-CoV-2 Research Assay (for Illumina) | A186XV6 | 16 rxn | M62111 |
| 96 rxn | M62112 | ||
| 960 rxn | M62113 | ||
| MultipSeq® SARS-CoV-2 Research Assay (for MGI DI) | A186XV7 | 16 rxn | M62121 |
| 96 rxn | M62122 | ||
| 960 rxn | M62123 |





