AIExome® Human Exome Panel V3 – Inherit is a version of AIExome® V3 specifically designed to detect rare and inherited diseases. In addition to the superior coverage of CDS region as AIExome® V3, AIExome® V3 – Inherit also covers clinically relevant non-coding pathogenic and likely pathogenic variants in ClinVar database, full length of human mitochondria genome and whole genome SNP backbone for CNV analysis to ensure the comprehensive understanding of rare and inherited diseases.
Panel Number | T600V1G |
Technical Platform | TargetSeq® Hybridization Capture Sequencing |
Coverage Size | 36.6 Mb |
Reference Database | RefSeq、MANE、CCDS、ClinVar、ClinGen etc. |
Reference Genome | GRCh38 (hg38) |
Coverage | CDS, ClinVar important SNPs, CNV backbone, mtDNA full-length |
Storage | <-70℃ |
Sequence Platform | Illumina / MGI |
Recommend Sequencing Read Length | PE150 |
Recommend Sequencing Data Size and Depth | 11Gb / 100X |
The AIExome® Human Exome Panel V3 product is available in 3 versions:
| CDS Region | Internal Reference QC SNPs | ClinVar | Mitochondria | SNP Backbone | Fusion | HLA | 641 Tumor-related Hotspot Genes | |
AIExome® V3 | ✓ | ✓ | ||||||
AIExome® V3 Inherit | ✓ | ✓ | ✓ | ✓ | ✓ | |||
AIExome® V3 Tomur | ✓ | ✓ | ✓ | ✓ | ✓ |
Covering over 99.9% of the CDS regions in major genetic databases.
High coverage of clinically relevant non-coding pathogenic and likely pathogenic variants in ClinVar database.
Complete capture of the full length of the mitochondrial genome.
Upgraded TargetSeq One® v2.0 kits and workflow for easier handling and more robust experiment. Compatible with automated workflow to reduce manual operation and human errors.
Stable data reproducibility across different NGS platforms, with excellent data performance and outstanding cost-efficiency.
Test results of AIExome® V3 – Inherit using NA12878 sample showed high sensitivity and accuracy in SNVs and InDels calling, suggesting AIExome® V3 – Inherit is a reliable panel for SNVs and InDels detection.
Table. Statistics of AIExome® V3 - Inherit using NA12878
| SNVs | InDels | |
Common Variants Detected | 17415 | 340 |
Sensitivity | 99.51% | 99.13% |
Accuracy | 99.41% | 92.64% |
Consistency | 98.92% | 91.89% |
AIExome® V3 – Inherit has a SNP backbone with probe density of 1 high MAF SNP per 100 kb at whole genome level, ensuring the large CNV (>500 kb) calling. At the same time, important CNV regions based on ClinGen database are covered with higher probe density of 1 high MAF SNP per 10 kb, and 1 high MAF SNP per 3 kb in the 15 kb of exon adjacent regions. AIExome® V3 - Inherit showed excellent performance of CNV calling in the comparison of CNV calling with major competitors.
Table. Comparison of CNV calling between AIExome® V3-Inherit and major competitors using NA12878
| chr14 | chr15 | chr16 | chr17 | chr22 | |
| Start | 105,740,061 | 30,172,500 | 35,213,828 | 46,111,085 | 22,037,881 |
| End | 105,862,093 | 30,407,000 | 35,530,832 | 46,215,376 | 22,916,769 |
| Length | 122,032 | 234,500 | 317,004 | 104,291 | 878,888 |
| Variation Type | del | del | dup | dup | del |
| WGS | Detected | Detected | Detected | Detected | Detected |
| Affy 750K | Detected | Not detected | Detected | Detected | Detected |
| AIExome® V3 - Inherit | Detected | Detected | Detected | Detected | Detected |
| Resolution | 500 kb | 50 kb | 500 kb | 50 kb | 500 kb |
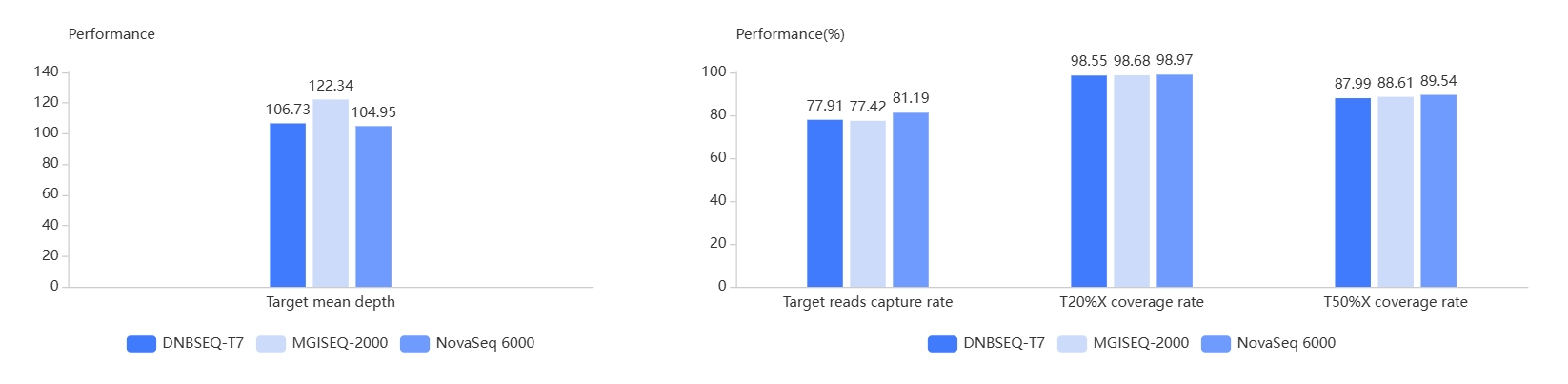
With newly upgraded TargetSeq One® v2.0 reagent kits and workflow, AIExome® V3 is not only simple and robust in workflow, but also excellent and consistent in performance across NGS platforms. The following figures show the performance of 10 Gb randomly selected raw sequencing data.
| Product Name | Panel Number | Set | Cat. No |
| AIExome® Human Exome Panel V3 - Inherit | T600V1G | 16 rxn | PT1008101 |
| 96 rxn | PT1008102 |

