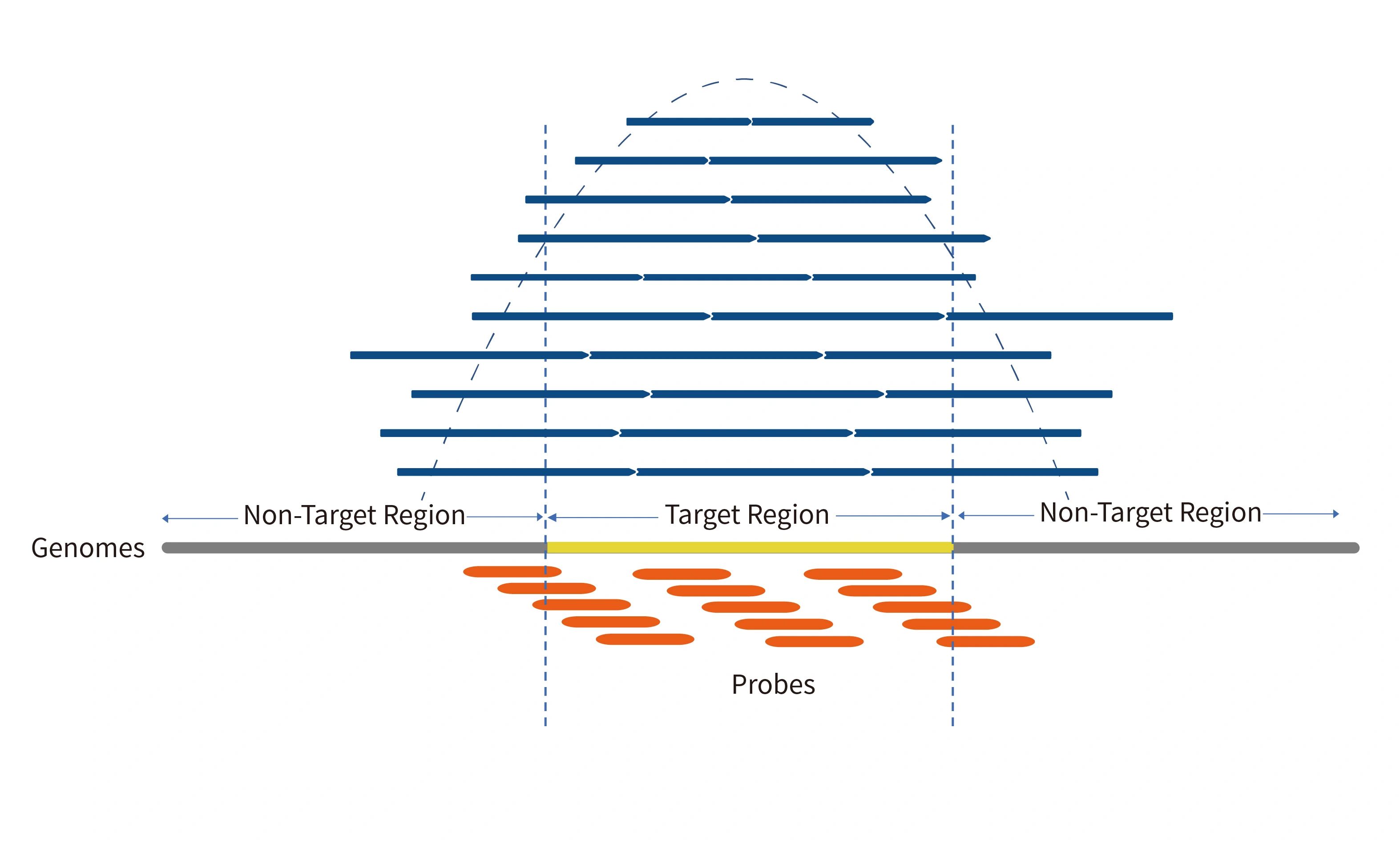
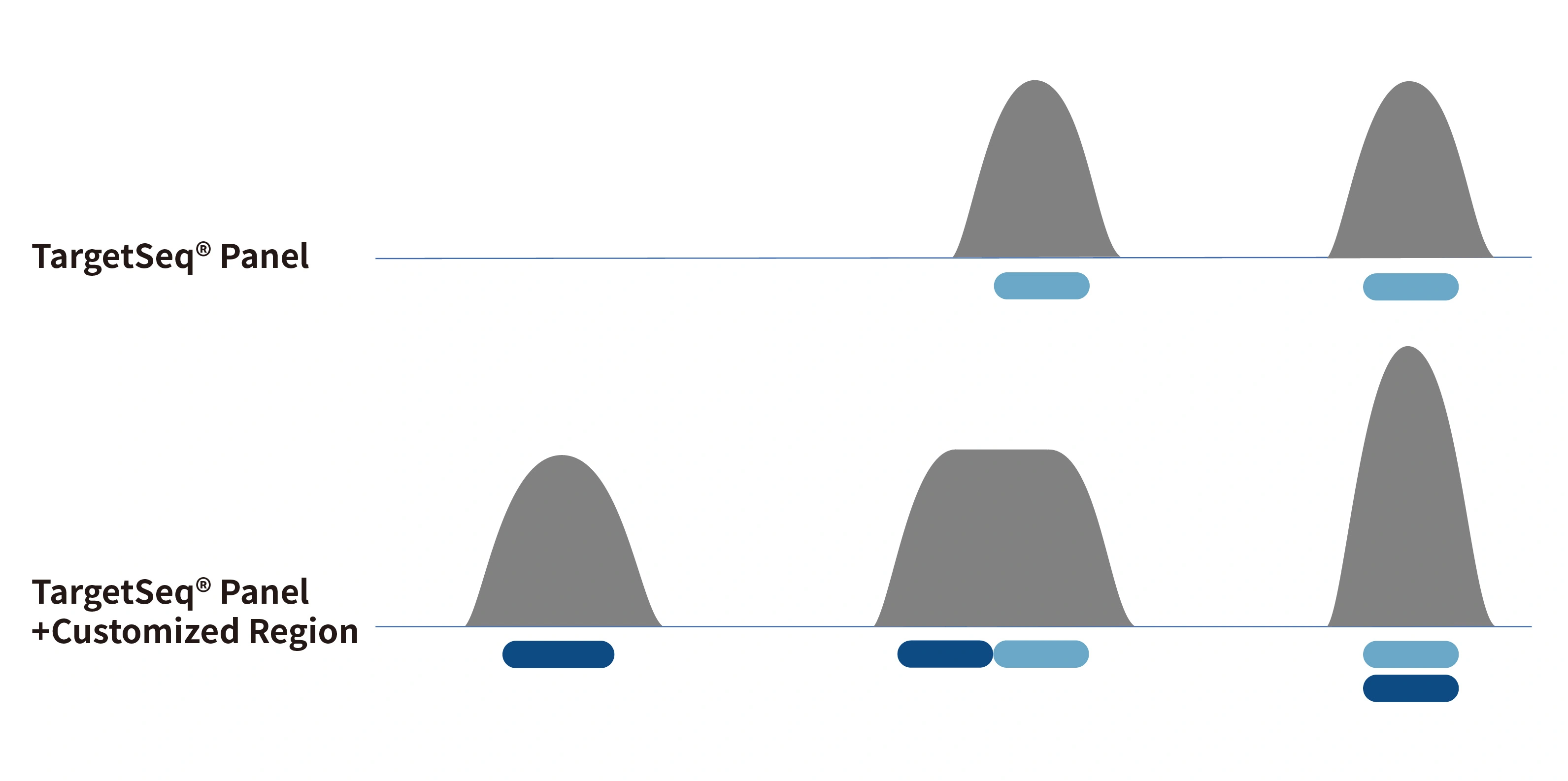
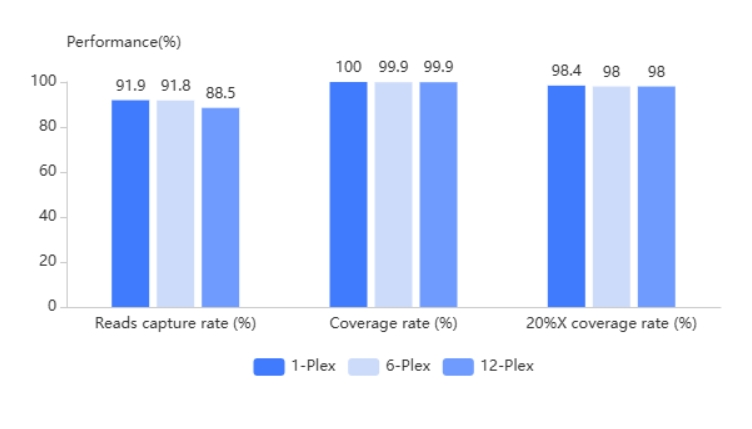
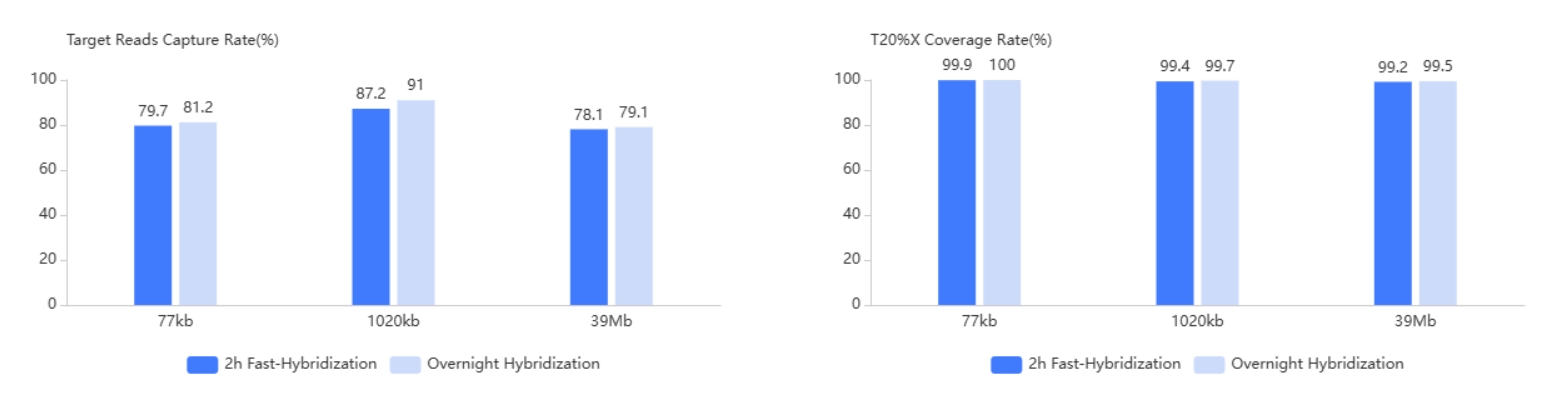
3× Tiling Probe Design
The TargetSeq®️ probe is designed with 3× tiling design scheme based on thermodynamic stability to achieve better coverage of target regions. At the same time, there are complete design schemes for reference genome fix or novel patches, hard-to-capture regions, different variant types, CNV, etc.

 CN
CN