Overview
TargetSeq® Solid Tumor Mid Panel covers 122 gene coding regions related to solid tumors, 10 hotspot fusion gene intron regions, 19 classic microsatellite sites, and 219 chemotherapy-related sites.
TargetSeq® Solid Tumor Mid Panel covers 122 gene coding regions related to solid tumors, 10 hotspot fusion gene intron regions, 19 classic microsatellite sites, and 219 chemotherapy-related sites.
Panel Number | T1288V1 |
Technical Platform | TargetSeq® Hybridization Capture Sequencing |
Coverage Size | 438.3 kb |
Reference Database | RefSeq |
Reference Genome | GRCh37 (hg19) |
Coverage | CDS region of 122 genes, 10 hotspot fusion gene intron regions, 19 MSI and 219 chemotherapy-related sites. |
Storage | -20℃±5℃ |
Sequence Platform | Illumina / MGI |
Recommend Sequencing Read Length | PE150 |
Recommend Sequencing Data Size and Depth | 1.5Gb/1,000X |
Extraordinary panel performance in coverage rate, capture rate and uniformity.
Proven sensitivity, specificity and reproducibility.
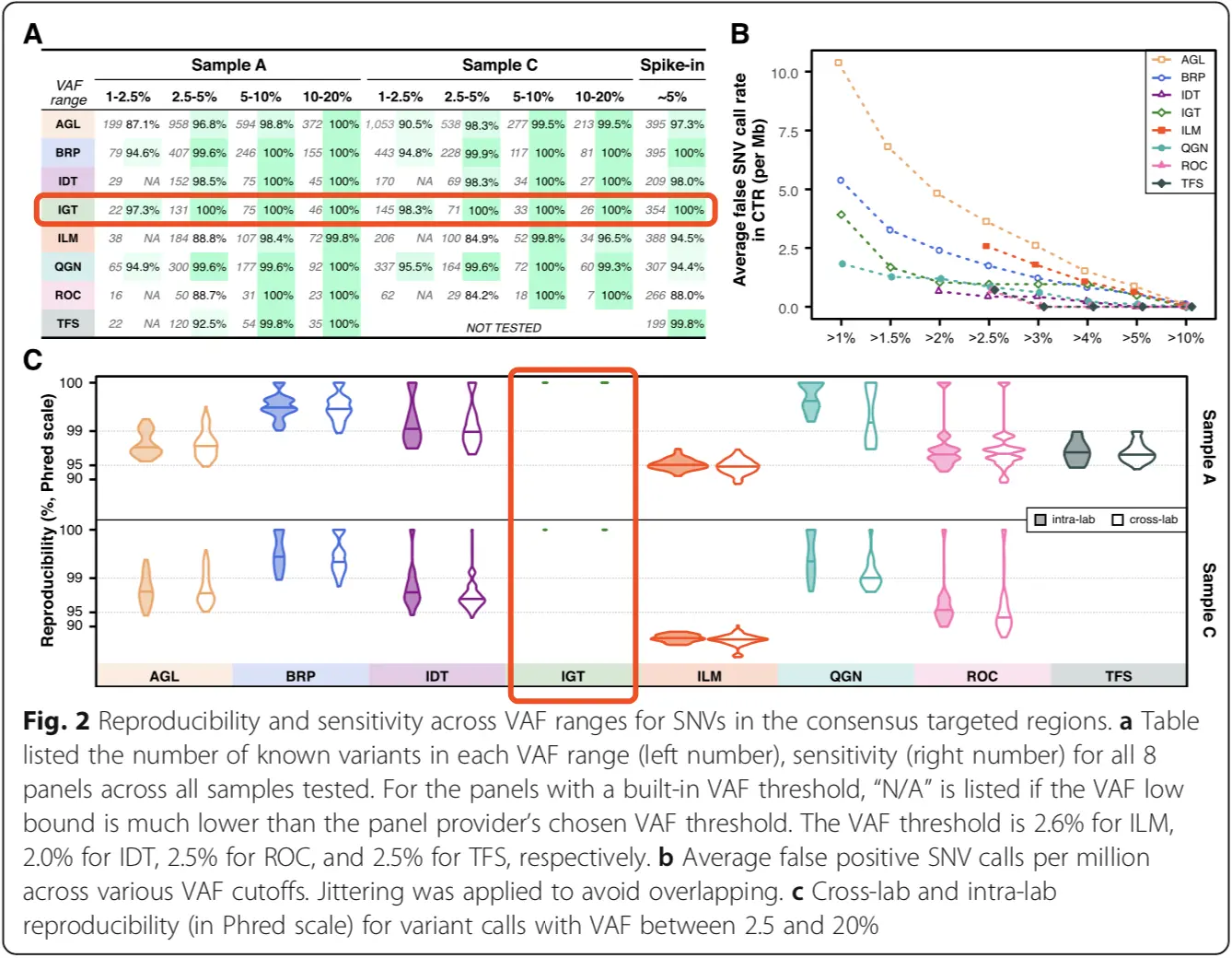
During an FDA-lead cross-platform multi-lab evaluation of eight Pan-Cancer panels, iGeneTech's TargetSeq® NCC Panel outperformed in sensitivity, specificity and reproducibility.
High sensitivity: >97% sensitivity at VAF of 1-2.5% and 100% sensitivity at VAF >2.5%
High specificity: 1/Mb false positive rate at VAF >5% and no false positive mutations detected at VAF >10%
High reproducibility: high reproducibility in cross-lab and intra-lab tests.
| Product Name | Panel Number | Set | Cat. No |
TargetSeq® Solid Tumor Mid Panel | T1288V1 | 16 rxn | PH2002101 |
| 96 rxn | PH2002102 |

