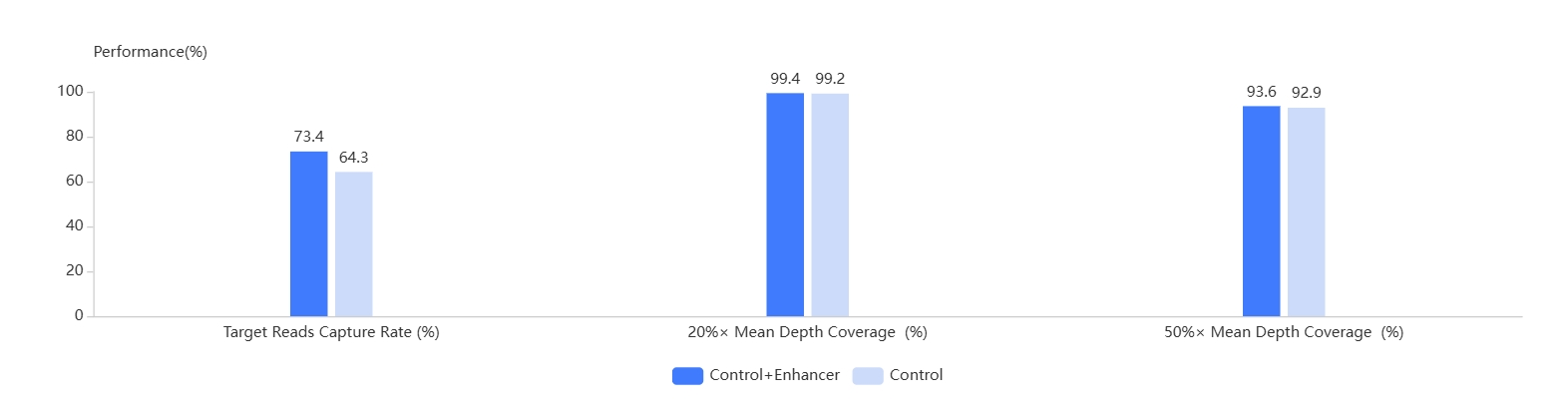
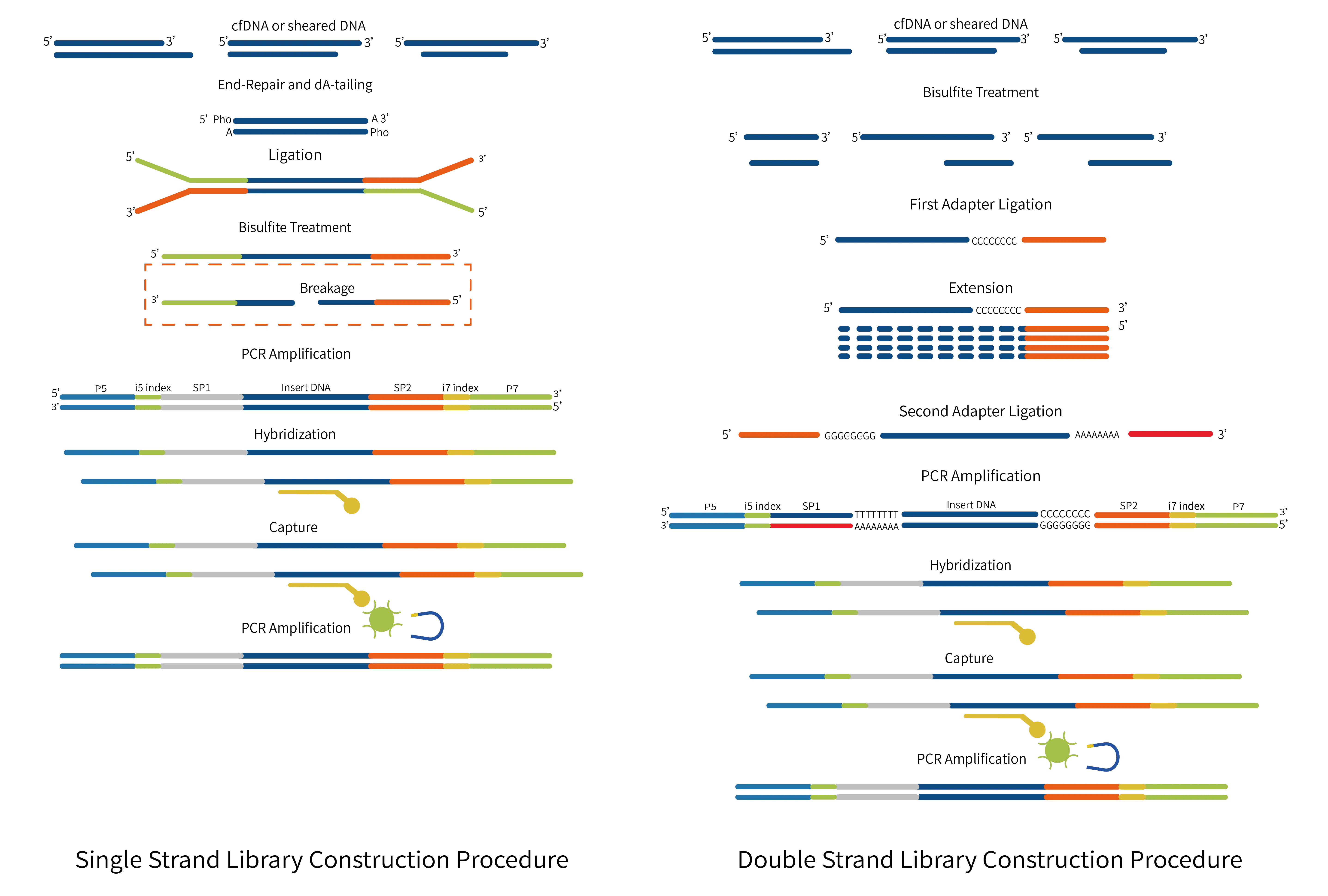
- AIExome® Human Exome Panel V3
- AIExome® Human Exome Panel V3-Inherited
- AIExome® Human Exome Panel V3-Tumor
- SARS-CoV-2 Panel
- HBV Panel
- EBV Panel
- Respiratory Pathogens Panel
- Gastrointestinal Pathogens Panel
- Oncogenic Pathogens Panel
- Immune-targeting pathogens Panel
- Vector-borne Pathogens Panel
- Animal Epizootic Pathogens Panel
- Zoonotic Pathogens Panel
- Mixed Multiple Pathogens Panel
- Target Sequencing Solution for Agriculture
- Animal Whole Exome Panel
- Multi-species Mitochondrial Panel
- Plant Whole Exome Panel
- MultipSeq® BCR-ABL1 Primer Pool
- MultipSeq® BRCA1/2 Research Assay V2
- MultipSeq® FLT3-ITD Primer Pool
- MultipSeq® TP53 Primer Pool
- TargetSeq® Core Genes Fusion RNA Panel
- TargetSeq® Lymphoma SV Panel
- TargetSeq® NSCLC Panel V2
- TargetSeq® Solid Tumor Fusion RNA Panel
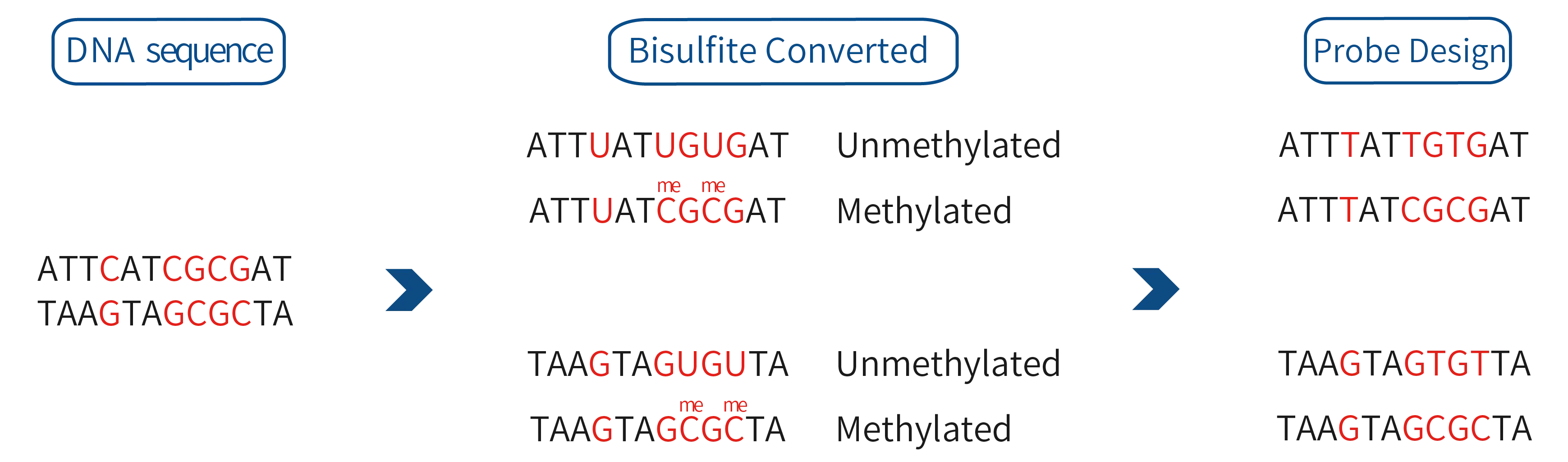
- BisCap® CpG Galaxy Panel
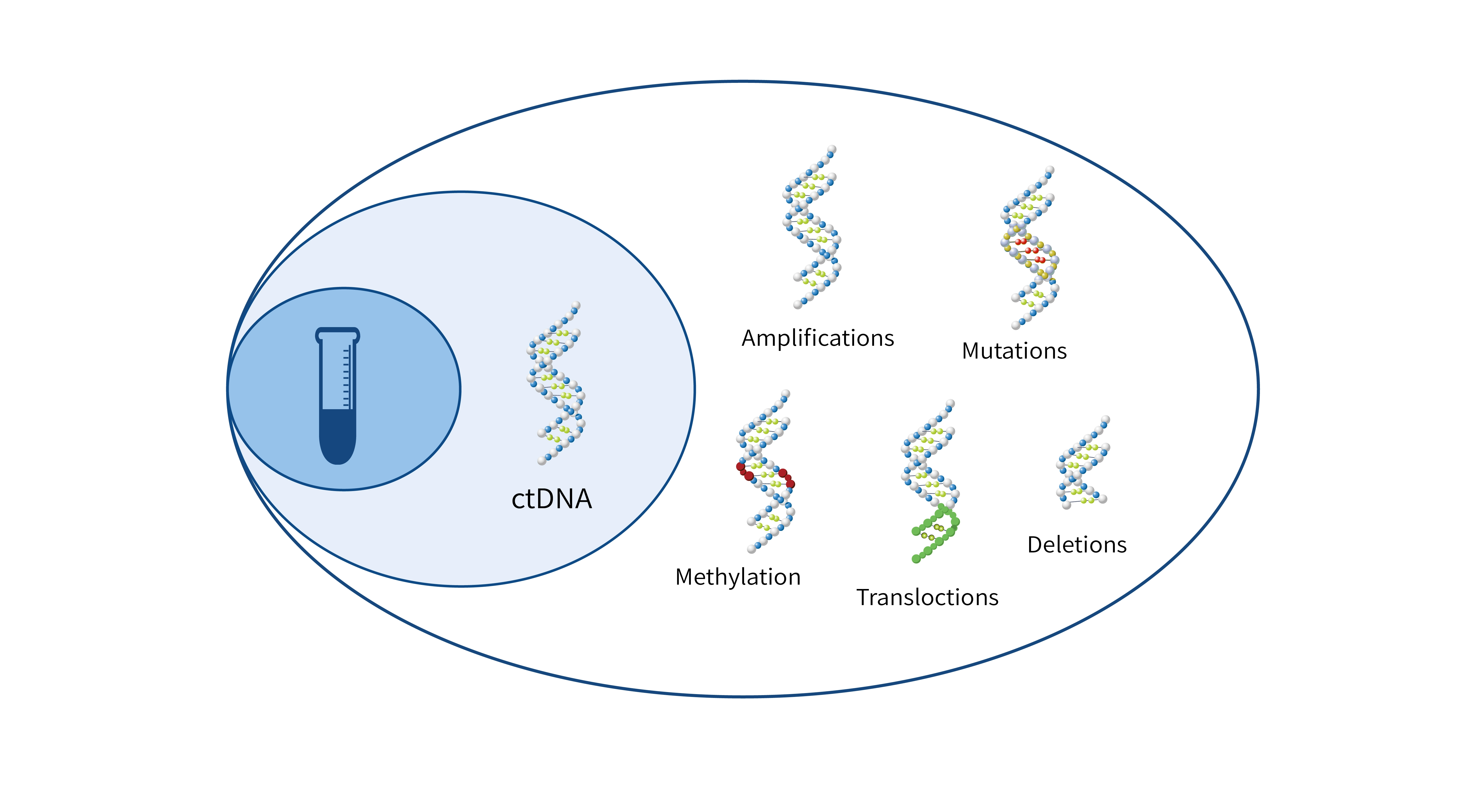
- BisCap® Multi-Cancer Early Detection Panel
- Pan-Cancer Panel
- TargetSeq® Solid Tumor Mid Panel
- Hema Tumor Fusion Panel
- CpG Island Methylation Panel
- HRD Panel & HRR Panel
EN
 CN
CN