MultipSeq® multiplex amplicon sequencing technology combines multiplex PCR with NGS technology. Based on the thermodynamic stability algorithm for the specific primer design at the genome-wide level and combined with the actual panel optimization process, MultipSeq® panels allow up to 5000-plex in a single tube with starting gDNA amount as low as 100 pg, which can quickly and efficiently achieve the enrichment of target regions for sequencing.
MultipSeq® multiplex amplicon sequencing technology uses multiplex PCR reactions to simultaneously amplify multiple target regions to obtain amplicon products, and then introduces the adapters to both sides of the amplicon products to obtain a specific amplicon library, which can be sequenced by using NGS platforms such as Illumina and MGI.
With self-developed professional quality control software, primers are designed under the guidance of internationally renowned experts with deep technical background.
Over 800 customizd MultipSeq® panels development experience, covering multiple application fields such as tumor companion diagnosis, drug evaluation, genetic screening, molecular breeding, and animal and plant evolution.
Suitable for target region sizes of <1 Mb. Can detect SNV, SNP, InDel, known fusions and other variants in the target region.
High compatibility and good consistency of test results with different starting amounts of samples to achieve consistent success rate under different conditions.
Optimization of key product metrics prior to Panel delivery for good performance of site coverage rate and uniformity.
Accurate detection of mutation frequency greater than 1% with good repeatability.
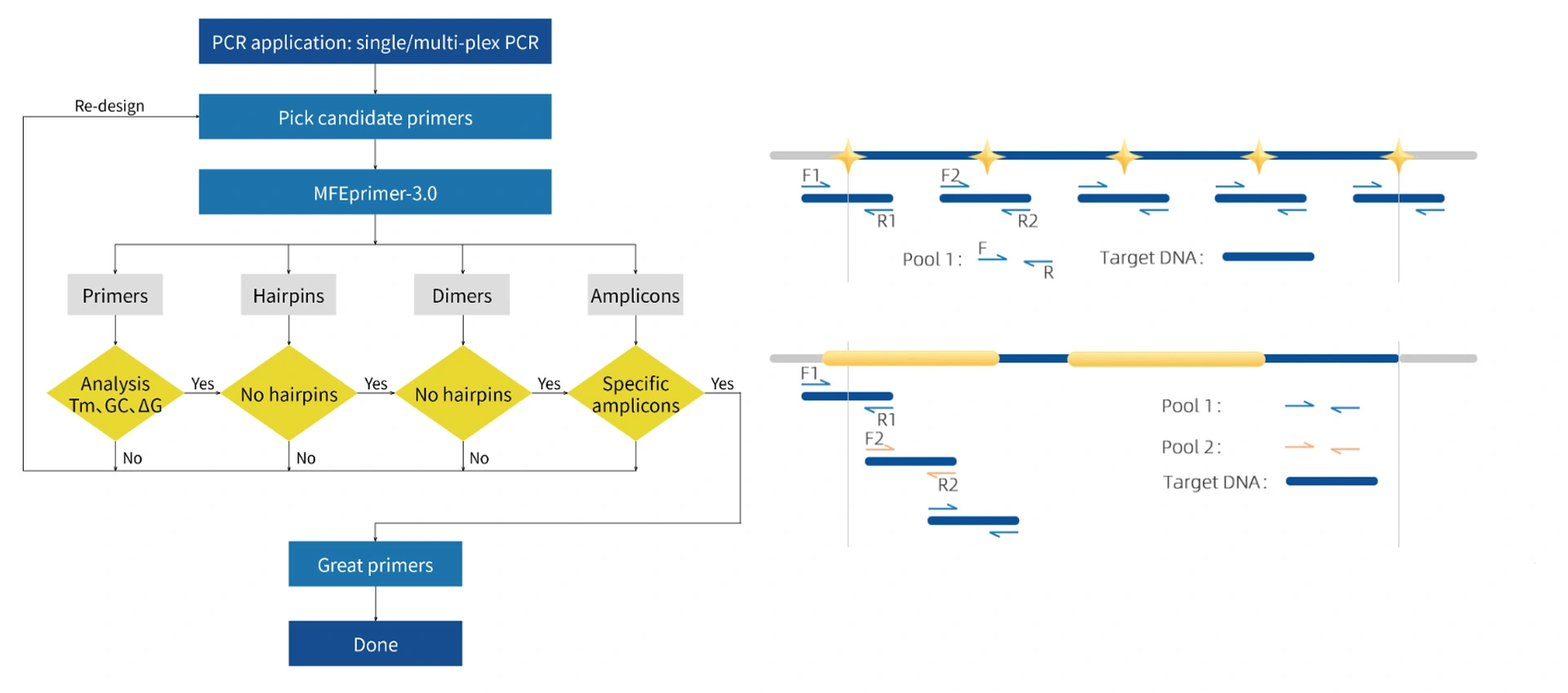
Self-developed and proven AIdesign primer design software and MFEprimer primer QC software are used in MultipSeq® panel design.
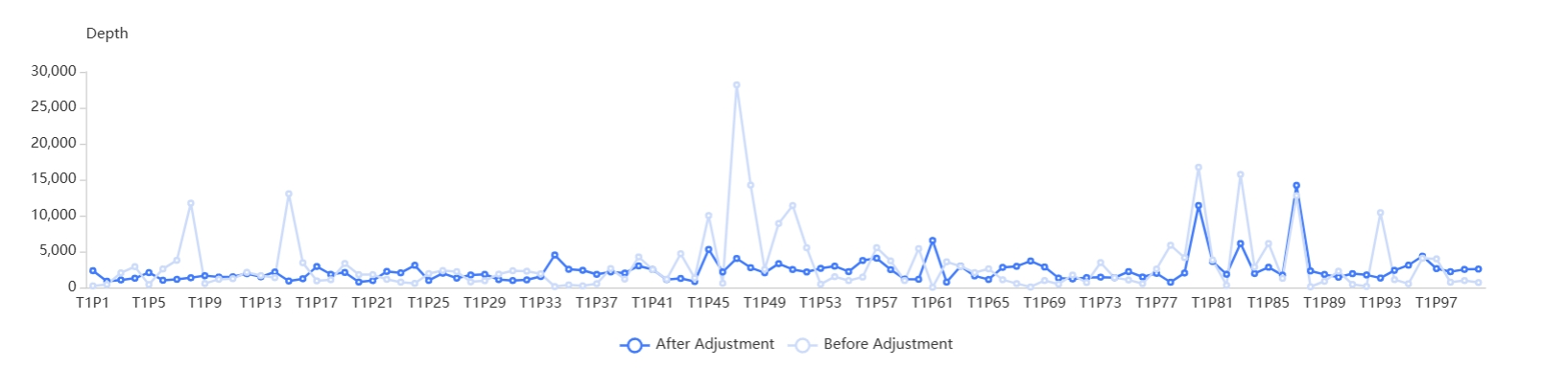
Before delivery, the key panel metrics will be tested and optimized to reduce amplification failure caused by high partiality, hard-to-cover regions or other reasons. And uniformity is optimized according to deep learning algorithms to guarantee product performance.
MultipSeq® multiplex amplicons sequencing technology is applicable to human, plant and animal, microbial, and other species with known genome. It is suitable for target capture of region sizes of <1 Mb. MultipSeq® panels are widely used in application fields such as oncology, genetic disorders, DTC genetic tests, immunology, microbiology, agriculture, and forensic science.

iGeneTech Bioscience provides both catalog and customized panels with reagent kits and automation upgrade solutions for the whole MultipSeq® workflow.
| Library Preparation Kits | Specific Primers | Equipments |
For continuous regions, our design strategy for primers is tiling and the amplicons are overlapping on each other to avoid the gap regions between amplicons. In this case, in order to avoid generating unwanted amplicons, primers of adjacent amplicons will be put into different pools. And for special hard-to-amplify regions like high GC content regions, an extra pool might be needed to ensure the performance.
There are some regions in the genome that we classify as hard-to-capture regions, including high GC content sequences, repetitive sequences and homologous sequences. Based on the evaluation result, some of these target regions might be dropped as they might affect the overall performance or introduce errors.
The lengths of the amplicons are affected by the sequencing read length, integrity of gDNA and application scenarios. Normally, the lengths of amplicons in MultipSeq® panels are 160 to 260 bp for PE150 sequencing, while they are shorter for PPFE or cfDNA panels but longer for STRs.